













| General Information: |
|
| Name(s) found: |
SIR_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 642 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] stromule [IDA] membrane [IDA] apoplast [IDA] chloroplast envelope [IDA] plastid [TAS] |
| Biological Process: |
response to cold
[IEP]
sulfate reduction [TAS] response to salt stress [IEP] |
| Molecular Function: |
sulfite reductase (ferredoxin) activity
[ISS]
copper ion binding [IDA] sulfite reductase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSTFRAPAG AATVFTADQK IRLGRLDALR SSHSVFLGRY GRGGVPVPPS ASSSSSSPIQ 60
61 AVSTPAKPET ATKRSKVEII KEKSNFIRYP LNEELLTEAP NVNESAVQLI KFHGSYQQYN 120
121 REERGGRSYS FMLRTKNPSG KVPNQLYLTM DDLADEFGIG TLRLTTRQTF QLHGVLKQNL 180
181 KTVMSSIIKN MGSTLGACGD LNRNVLAPAA PYVKKDYLFA QETADNIAAL LSPQSGFYYD 240
241 MWVDGEQFMT AEPPEVVKAR NDNSHGTNFV DSPEPIYGTQ FLPRKFKVAV TVPTDNSVDL 300
301 LTNDIGVVVV SDENGEPQGF NIYVGGGMGR THRMESTFAR LAEPIGYVPK EDILYAVKAI 360
361 VVTQREHGRR DDRKYSRMKY LISSWGIEKF RDVVEQYYGK KFEPSRELPE WEFKSYLGWH 420
421 EQGDGAWFCG LHVDSGRVGG IMKKTLREVI EKYKIDVRIT PNQNIVLCDI KTEWKRPITT 480
481 VLAQAGLLQP EFVDPLNQTA MACPAFPLCP LAITEAERGI PSILKRVRAM FEKVGLDYDE 540
541 SVVIRVTGCP NGCARPYMAE LGLVGDGPNS YQVWLGGTPN LTQIARSFMD KVKVHDLEKV 600
601 CEPLFYHWKL ERQTKESFGE YTTRMGFEKL KELIDTYKGV SQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
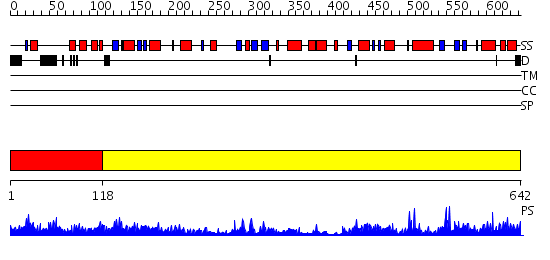
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..117] | 1.2 | Structure of Mycobacterium tuberculosis NirA protein | |
| 2 | View Details | [118..642] | 95.69897 | SULFITE REDUCTASE STRUCTURE AT 1.6 ANGSTROM RESOLUTION |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.71 |
Source: Reynolds et al. (2008)