













| General Information: |
|
| Name(s) found: |
CYP38_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 437 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] thylakoid lumen [IDA] chloroplast thylakoid lumen [IDA][ISS] thylakoid [IDA] chloroplast thylakoid membrane [IDA] |
| Biological Process: |
protein folding
[ISS]
defense response to bacterium [IEP] |
| Molecular Function: |
peptidyl-prolyl cis-trans isomerase activity
[IEA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAAFASLPT FSVVNSSRFP RRRIGFSCSK KPLEVRCSSG NTRYTKQRGA FTSLKECAIS 60
61 LALSVGLMVS VPSIALPPNA HAVANPVIPD VSVLISGPPI KDPEALLRYA LPIDNKAIRE 120
121 VQKPLEDITD SLKIAGVKAL DSVERNVRQA SRTLQQGKSI IVAGFAESKK DHGNEMIEKL 180
181 EAGMQDMLKI VEDRKRDAVA PKQKEILKYV GGIEEDMVDG FPYEVPEEYR NMPLLKGRAS 240
241 VDMKVKIKDN PNIEDCVFRI VLDGYNAPVT AGNFVDLVER HFYDGMEIQR SDGFVVQTGD 300
301 PEGPAEGFID PSTEKTRTVP LEIMVTGEKT PFYGSTLEEL GLYKAQVVIP FNAFGTMAMA 360
361 REEFENDSGS SQVFWLLKES ELTPSNSNIL DGRYAVFGYV TDNEDFLADL KVGDVIESIQ 420
421 VVSGLENLAN PSYKIAG |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
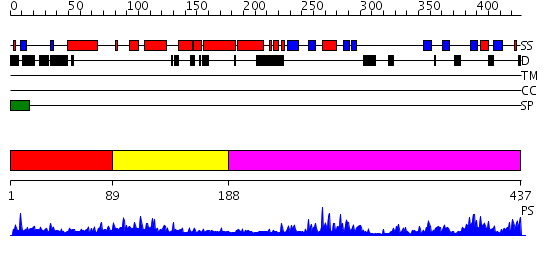
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..88] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [89..187] | 7.69897 | Solution Structure of the RNA recognition motif in Peptidyl-prolyl cis-trans isomerase E | |
| 3 | View Details | [188..437] | 38.0 | Crystal Structure of cyclophilin from Plasmodium yoelii. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|