













| General Information: |
|
| Name(s) found: |
AMPD_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 839 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
microsome [IDA] cytosol [IDA] |
| Biological Process: |
embryonic development ending in seed dormancy
[IMP]
purine ribonucleoside monophosphate biosynthetic process [ISS] |
| Molecular Function: |
AMP deaminase activity
[IGI][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEPNIYQLAL AALFGASFVA VSGFFMHFKA LNLVLERGKE RKENPDGDEP QNPTLVRRRS 60
61 QVRRKVNDQY GRSPASLPDA TPFTDGGGGG GGDTGRSNGH VYVDEIPPGL PRLHTPSEGR 120
121 ASVHGASSIR KTGSFVRPIS PKSPVASASA FESVEESDDD DNLTNSEGLD ASYLQANGDN 180
181 EMPADANEEQ ISMAASSMIR SHSVSGDLHG VQPDPIAADI LRKEPEQETF VRLNVPLEVP 240
241 TSDEVEAYKC LQECLELRKR YVFQETVAPW EKEVISDPST PKPNTEPFAH YPQGKSDHCF 300
301 EMQDGVVHVF ANKDAKEDLF PVADATAFFT DLHHVLKVIA AGNIRTLCHR RLVLLEQKFN 360
361 LHLMLNADKE FLAQKSAPHR DFYNVRKVDT HVHHSACMNQ KHLLRFIKSK LRKEPDEVVI 420
421 FRDGTYLTLR EVFESLDLTG YDLNVDLLDV HADKSTFHRF DKFNLKYNPC GQSRLREIFL 480
481 KQDNLIQGRF LGEITKQVFS DLEASKYQMA EYRISIYGRK MSEWDQLASW IVNNDLYSEN 540
541 VVWLIQLPRL YNIYKDMGIV TSFQNILDNI FIPLFEATVD PDSHPQLHVF LKQVVGFDLV 600
601 DDESKPERRP TKHMPTPAQW TNAFNPAFSY YVYYCYANLY VLNKLRESKG MTTITLRPHS 660
661 GEAGDIDHLA ATFLTCHSIA HGINLRKSPV LQYLYYLAQI GLAMSPLSNN SLFLDYHRNP 720
721 FPVFFLRGLN VSLSTDDPLQ IHLTKEPLVE EYSIAASVWK LSACDLCEIA RNSVYQSGFS 780
781 HALKSHWIGK DYYKRGPDGN DIHKTNVPHI RVEFRDTIWK EEMQQVYLGK AVISDEVVP |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
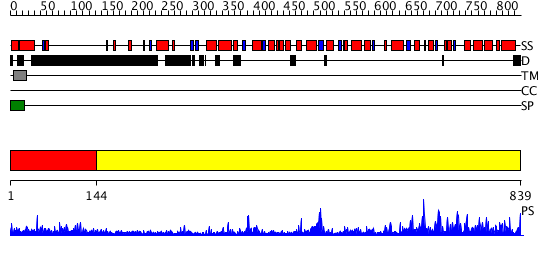
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..143] | 3.39794 | No description for 1y0fB was found. | |
| 2 | View Details | [144..839] | 1000.0 | X-Ray Structure of Adenosine 5'-Monophosphate Deaminase from Arabidopsis Thaliana in Complex with Coformycin 5'-Phosphate |
| Source: Reynolds et al. 2008. Manuscript submitted |

|