













| General Information: |
|
| Name(s) found: |
TPIC_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 315 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA][ISS] mitochondrion [IDA] apoplast [IDA] chloroplast envelope [IDA] thylakoid [IDA] |
| Biological Process: |
multicellular organism reproduction
[IMP]
primary root development [IMP] reductive pentose-phosphate cycle [ISS] glyceraldehyde-3-phosphate biosynthetic process [IDA] glycerol catabolic process [IMP] chloroplast organization [IMP] triglyceride mobilization [IMP] |
| Molecular Function: |
triose-phosphate isomerase activity
[IDA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAATSLTAPP SFSGLRRISP KLDAAAVSSH QSFFHRVNSS TRLVSSSSSS HRSPRGVVAM 60
61 AGSGKFFVGG NWKCNGTKDS IAKLISDLNS ATLEADVDVV VSPPFVYIDQ VKSSLTDRID 120
121 ISGQNSWVGK GGAFTGEISV EQLKDLGCKW VILGHSERRH VIGEKDEFIG KKAAYALSEG 180
181 LGVIACIGEK LEEREAGKTF DVCFAQLKAF ADAVPSWDNI VVAYEPVWAI GTGKVASPQQ 240
241 AQEVHVAVRG WLKKNVSEEV ASKTRIIYGG SVNGGNSAEL AKEEDIDGFL VGGASLKGPE 300
301 FATIVNSVTS KKVAA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
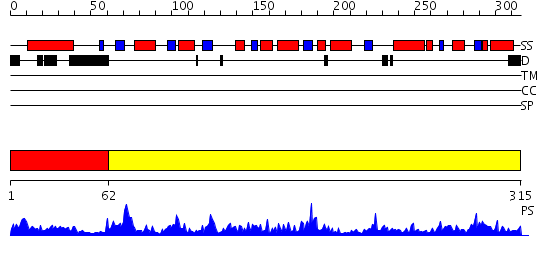
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..61] | 11.522879 | No description for 1zmrA was found. | |
| 2 | View Details | [62..315] | 96.154902 | CRYSTAL STRUCTURE OF RABBIT MUSCLE TRIOSEPHOSPHATE ISOMERASE |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)