













| General Information: |
|
| Name(s) found: |
GLGL3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 521 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
starch biosynthetic process
[TAS]
|
| Molecular Function: |
glucose-1-phosphate adenylyltransferase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSCCNFSLG TKTVLAKDSF KNVENKFLGE KIKGSVLKPF SSDLSSKKFR NRKLRPGVAY 60
61 AIATSKNAKE ALKNQPSMFE RRRADPKNVA AIILGGGDGA KLFPLTKRAA TPAVPVGGCY 120
121 RMIDIPMSNC INSCINKIFV LTQFNSASLN RHLARTYFGN GINFGDGFVE VLAATQTPGE 180
181 AGKKWFQGTA DAVRKFLWVF EDAKNRNIEN IIILSGDHLY RMNYMDFVQH HVDSKADITL 240
241 SCAPVDESRA SEYGLVNIDR SGRVVHFSEK PTGIDLKSMQ TDTTMHGLSH QEAAKSPYIA 300
301 SMGVYCFKTE ALLKLLTWRY PSSNDFGSEI IPAAIKDHNV QGYIYRDYWE DIGTIKSFYE 360
361 ANIALVEEHP KFEFYDQNTP FYTSPRFLPP TKTEKCRIVN SVISHGCFLG ECSIQRSIIG 420
421 ERSRLDYGVE LQDTLMLGAD SYQTESEIAS LLAEGNVPIG IGRDTKIRKC IIDKNAKIGK 480
481 NVVIMNKDDV KEADRPEEGF YIRSGITVVV EKATIKDGTV I |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
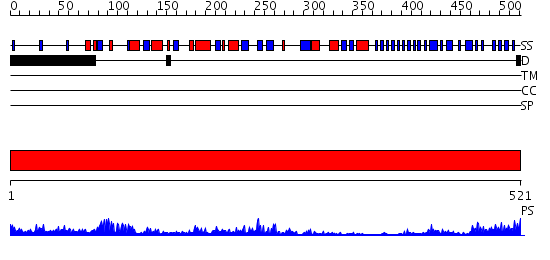
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..521] | 79.69897 | Crystal structure of potato tuber ADP-glucose pyrophosphorylase |
Functions predicted (by domain):
| # | Gene Ontology predictions |
| 1 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)