













| General Information: |
|
| Name(s) found: |
CE34275
[WormBase]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 557 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA chromatin [IEA |
| Biological Process: |
hermaphrodite genitalia development
[IMP positive regulation of growth rate [IMP chromatin assembly or disassembly [IEA gamete generation [IMP |
| Molecular Function: |
chromatin binding
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQSTSSEGS GPAAKIARKN SPENDPETAE MFIVDEITDM RYSTTCRQLQ FLVKWAIPGS 60
61 EMTWQPLDDF PFGLNHFSIL DYRKDHEEKY EACLQKQRRS KVMKDDDRSW NVSRNSNRSR 120
121 NRSKSSSKPA QRTEPMTQRE KFDAILQISE ISLEKQYKLT NKKVPRRSQF TDPDAPKKKS 180
181 KGARHYRDQN LKTLFKSSLI RKYRKPQESD DDLEPDVAQD SPENTPAPPT KSPESSGFSE 240
241 NLENLGANTP SPSPDFEEEH LEKKPETGNL TENRPKRVYS HETAESMENY AKLLLYTAFD 300
301 TKSWETLGGH IKNGFRLSFV CSNGLTIIEN VVELNKNLNK FSDSRESRMV ADLFELLLKK 360
361 GVEFDEKTLP PTMCAAVRST RYDIRRDFRC LHNQYIDKRP RMCEWFSATG GGGSCGTTEI 420
421 CTFWNNDAGY LPNTHQRSKF HVDLSEFTGN NLGLLAFSMA WHEDFIDNNS GGPEIVKNNR 480
481 FNAHIDVPCF TLWMNGKMCE KLENSHGLSI FRPLGTVLDV NELKIKITEH SSRFVAILPV 540
541 FNLNSDWKNC FFPLENQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
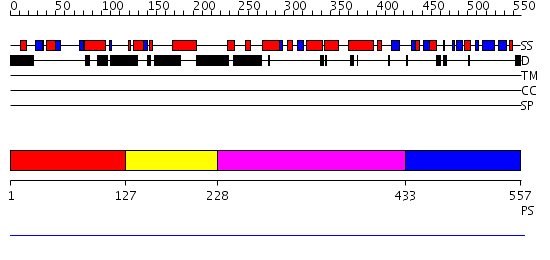
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..126] | 1.13 | Heterochromatin protein 1, HP1 | |
| 2 | View Details | [127..227] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [228..432] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [433..557] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)