













| General Information: |
|
| Name(s) found: |
pry-1 /
CE30125
[WormBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 586 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA |
| Biological Process: |
negative regulation of vulval development
[IGI embryonic cleavage [IMP spindle organization [IMP regulation of cell fate specification [IMP multicellular organismal development [IEA anterior/posterior pattern formation [IMP negative regulation of Wnt receptor signaling pathway [IGI cell migration [IMP |
| Molecular Function: |
signal transducer activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METHLGWARS LEAVLSDRSA LDAFQEWLIE YSSPQYLDLF FAIRAYERMA LEGKPEKSQL 60
61 SKSIYSKFLS SRTGNCEAIP KHFRAPIGEK LRHGTELEDR VFSHCSNFVQ EFLRRQHEEF 120
121 VGSEEFIEAF NKMSSTTADQ LPGGSAHHSS HQNTMRRSSG TTSRKSAAQI ATQLTAEALL 180
181 KSKHDRHSKL GETKLEKMYP PTRQPYVCNA TTSHNDSAVS STFSGDTPEA HRMHSNRLRH 240
241 IRDEQARENH GTMTLPRVEK ASVDGQQWDH SSESGRRNFA MEITRKLLRH IDKVKLNDEM 300
301 EKRIDDIEEC RYTTIDMVNG TEPNDDLGKI DEDEELDDYL KMKMTDDSQK GSTNRSPKGP 360
361 AGEPNKSGEG SKNTTLSPTN RAPAQLHNTI RVPRRKDYPR DTSASLKSHR HHQIDTNRMM 420
421 SQSMCAPSYS SASSSYSRDS FAPAPTTRVN FAPGSSKSSQ FYDSSGIGSM APSAFSATSS 480
481 LDYKDRRQHR KAPTPKKHSK IGKNLSNLIT ISYLGTDKIP VVTHVPNDGP MTLAEFKRHF 540
541 ALPNGAHQLF FKTECEDGSA PFQLLLIKDE HHLLPVFEGR IAAELR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
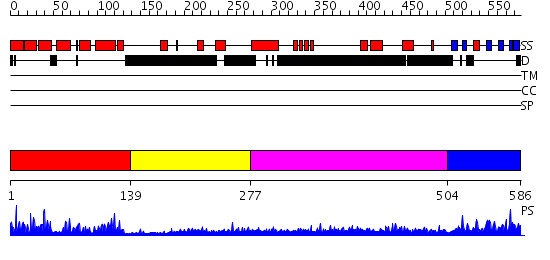
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..138] | 41.0 | No description for 2jm5A was found. | |
| 2 | View Details | [139..276] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [277..503] | 2.100987 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [504..586] | 28.0 | Crystal structure of axin dix domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)