













| General Information: |
|
| Name(s) found: |
hst2 /
SPCC132.02
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 332 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mating-type region heterochromatin
[IDA nucleus [IDA nuclear chromatin [IDA telomeric heterochromatin [IDA cytoplasm [IDA cytosol [IDA rDNA heterochromatin [IDA chromatin remodeling complex [ISS centromeric heterochromatin [IDA |
| Biological Process: |
chromatin remodeling
[ISS chromatin silencing at centromere [IMP chromatin silencing at rDNA [IMP histone deacetylation [TAS regulation of nucleosome density [IEP |
| Molecular Function: |
zinc ion binding
[IEA]
transcription corepressor activity [IEP transcription regulator activity [IC NAD or NADH binding [IEA] NAD-dependent histone deacetylase activity [TAS specific transcriptional repressor activity [IEP NAD binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVKNTVKHVD SSKHLEKVAS LIKEGKVKKI CVMVGAGIST AAGIPDFRSP ETGIYNNLQR 60
61 FNLPYAEAVF DLSYFRKNPR PFYELAHELM PEKYRPTYTH YFIRLLHDKR LLQKCYTQNI 120
121 DTLERLAGVP DKALIEAHGS FQYSRCIECY EMAETEYVRA CIMQKQVPKC NSCKGLIKPM 180
181 IVFYGEGLPM RFFEHMEKDT KVCDMALVIG TSLLVHPFAD LPEIVPNKCQ RVLINREPAG 240
241 DFGERKKDIM ILGDCDSQVR ALCKLLGWSD ELEKLIDTSV ETLTEEISLL SVDSTIEKNA 300
301 SEQKKDDNSV NPFTKIEEKK KDEVTLLVSD DE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
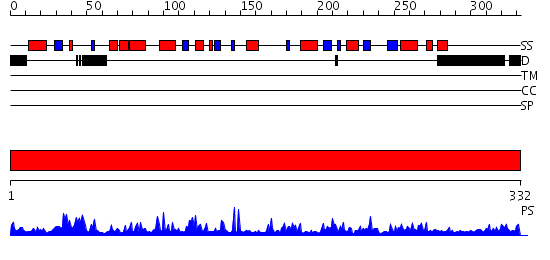
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..332] | 83.0 | Sirt2 histone deacetylase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)