













| General Information: |
|
| Name(s) found: |
SPBC1683.04
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 832 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
carbohydrate catabolic process
[IC cellular catabolic process [NAS] |
| Molecular Function: |
beta-glucosidase activity
[IEA]
hydrolase activity, hydrolyzing O-glycosyl compounds [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMEHDVEDLI NQLDISEKAM LLSGTDLWHT AAIPRLNIPS IRLSDGPNGI RGTSFFNSSP 60
61 SACFPCGTAL GATFDKKLLF EVGEYLAEEA KAKGVSVVLG PTVNIHRGPL NGRGFESFSE 120
121 DSTLSGLAAS YVILGLQSKN VQACIKHFVC NDMEDERNSV SIDVSQRALR EVYLMPFQLA 180
181 CKYSNFKSLM TSYNKVNGEH VSQSRILLDN ILRKEWEWKG TIISDWFGTY SLKKAIDAGL 240
241 DLEMPGKPRF RNVNTIQHLV GSKELSESIL DERAKNVLKL VKHSWQNTEA ENHCELNNDS 300
301 SCLREALKKF ASQSIVLLKN KKKLLPLSRK GTFAVIGPNA KVCNYSGGGS ANLKPYYTVS 360
361 MYDGIAAKID GVPEYALGCH NYLNLPNIAN LLVNPRTGKH GYVAKFYLEP ATSENRTLID 420
421 DYDLEDGVVR FYDYCNDKMK DGYFYIDIEG YLIPDEDAVY EFGISVFGTA LLFIDDVLLI 480
481 DNKTKQTPTN HTFEFGTIEE RNSIYLKKGR KYNVRVEYGS AATYTLSTNL SPSTGGRYSI 540
541 GCVKVIDPET EIDYAVRVAK SVDCVILCVG LTAEWETEGE DRKTMTLPSL SDKLVYSILQ 600
601 SNPNTVVVTQ SGTPIEMPWI SEAHTLLHIW YNGNELGNAL ANIIFGEQNP CGKLPITFPK 660
661 KLKDNPAYLS FRSSRGHCVY GEDVFVGYKY YEAVEREVLF PFGYGLSYTT FELSNLYLKN 720
721 CGERLRIDLE ISNTGPMSGA EIIQVYISQI VRSVNRPVKE LKEFSKVVLC PKETKLIRIE 780
781 LDIKYATSFY DELNEKWCSE EGEYNVLVGT SSKDIALTGK FTLPKTIHWT GL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
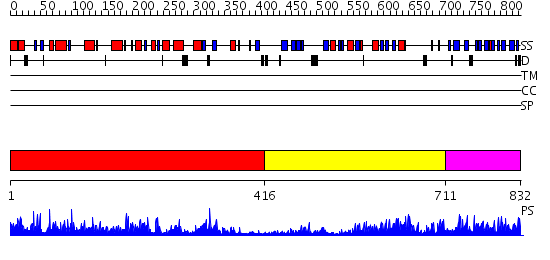
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..415] | 95.39794 | Beta-D-glucan exohydrolase, N-terminal domain; Beta-D-glucan exohydrolase, C-terminal domain | |
| 2 | View Details | [416..710] | 3.77 | Beta-D-glucan exohydrolase, N-terminal domain; Beta-D-glucan exohydrolase, C-terminal domain | |
| 3 | View Details | [711..832] | 3.06 | Dex49A from Penicillium minioluteum |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)