













| General Information: |
|
| Name(s) found: |
SPAC3H8.02
[Sanger Pombe]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 444 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[ISS |
| Biological Process: |
cellular cell wall organization
[ISS phospholipid transport [ISS |
| Molecular Function: |
phosphatidylinositol transporter activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPEGAGRPWN LTELEEEKLK TMWSYLFKLF GITLLERTES WYTVKTHLSD DSSSSSSHRL 60
61 SSVSYAKSRT RLELTSSSHG SDTRSFNDKT KNVHLERVEK IASEWDPEGL RVCFWDAVNC 120
121 DDPDGLLLRF LRARKWNVEA ALEMFMKTVH WRSREMNVGE IVCNADHLDK DDDFVRQLRI 180
181 GKCFIFGEDK HNRPVCYIRA RLHKVGDVSP ESVERLTVWV METARLILKP PIETATVVFD 240
241 MTDFSMSNMD YGPLKFMIKC FEAHYPECLG ECIVHKAPWL FQGVWSIIKS WLDPVVVSKV 300
301 KFTRNYRDLQ QYINPDNILK EFGGPNPWRY TYPEPCQNEA EALKNVEARK SLRAKKDAIA 360
361 KQYEEVTMDW ILNNGDMAEV KQKRRKLASQ LIDAYWNLDK YIRARSVYDR MGLIAPQTSH 420
421 TLLLSQPTNG DVKESMVEVT SSAT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
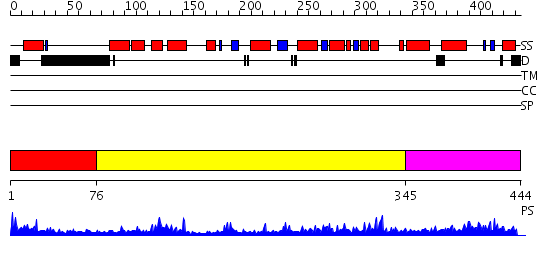
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..75] | 1.211996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [76..344] | 46.39794 | PHOSPHATIDYLINOSITOL TRANSFER PROTEIN SEC14P FROM SACCHAROMYCES CEREVISIAE | |
| 3 | View Details | [345..444] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)