













| General Information: |
|
| Name(s) found: |
ure2 /
SPAC1952.11c
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 835 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
urea metabolic process
[IEA]
nitrogen compound metabolic process [IEA] |
| Molecular Function: |
nickel ion binding
[IEA]
urease activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQPRELHKLT LHQLGSLAQK RLCRGVKLNK LEATSLIASQ IQEYVRDGNH SVADLMSLGK 60
61 DMLGKRHVQP NVVHLLHEIM IEATFPDGTY LITIHDPICT TDGNLEHALY GSFLPTPSQE 120
121 LFPLEEEKLY APENSPGFVE VLEGEIELLP NLPRTPIEVR NMGDRPIQVG SHYHFIETNE 180
181 KLCFDRSKAY GKRLDIPSGT AIRFEPGVMK IVNLIPIGGA KLIQGGNSLS KGVFDDSRTR 240
241 EIVDNLMKQG FMHQPESPLN MPLQSARPFV VPRKLYAVMY GPTTNDKIRL GDTNLIVRVE 300
301 KDFTEYGNES VFGGGKVIRD GTGQSSSKSM DECLDTVITN AVIIDHTGIY KADIGIKNGY 360
361 IVGIGKAGNP DTMDNIGENM VIGSSTDVIS AENKIVTYGG MDSHVHFICP QQIEEALASG 420
421 ITTMYGGGTG PSTGTNATTC TPNKDLIRSM LRSTDSYPMN IGLTGKGNDS GSSSLKEQIE 480
481 AGCSGLKLHE DWGSTPAAID SCLSVCDEYD VQCLIHTDTL NESSFVEGTF KAFKNRTIHT 540
541 YHVEGAGGGH APDIISLVQN PNILPSSTNP TRPFTTNTLD EELDMLMVCH HLSRNVPEDV 600
601 AFAESRIRAE TIAAEDILQD LGAISMISSD SQAMGRCGEV ISRTWKTAHK NKLQRGALPE 660
661 DEGSGVDNFR VKRYVSKYTI NPAITHGISH IVGSVEIGKF ADLVLWDFAD FGARPSMVLK 720
721 GGMIALASMG DPNGSIPTVS PLMSWQMFGA HDPERSIAFV SKASITSGVI ESYGLHKRVE 780
781 AVKSTRNIGK KDMVYNSYMP KMTVDPEAYT VTADGKVMEC EPVDKLPLSQ SYFIF |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
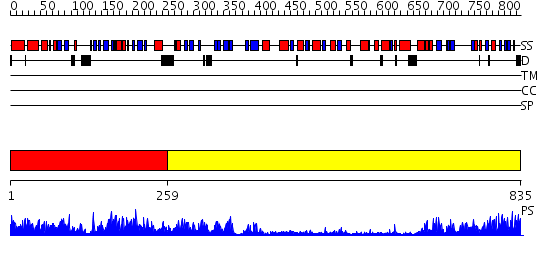
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..258] | 104.0 | Urease, beta-subunit; Urease, gamma-subunit | |
| 2 | View Details | [259..835] | 177.0 | alpha-Subunit of urease; alpha-subunit of urease, catalytic domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)