













| General Information: |
|
| Name(s) found: |
cut3 /
SPBC146.03c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1324 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA inner kinetochore of condensed chromosome [IGI cytoplasm [IDA cytosol [IDA nucleolus [IGI condensin complex [IDA |
| Biological Process: |
mitotic chromosome condensation
[IMP mitotic sister chromatid segregation [IMP cellular protein localization [ISS |
| Molecular Function: |
ATP binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDKGIFRTS STPSIVDVTP DRGERPRKLV RSVLESPSQK DVASIVKIEQ TPSRPFFNDF 60
61 LKKRITDSLN ERPNLLNKFM SAQDGTPSKS TGFNERSSQL VSEFTTTEDI ENCEETTQVL 120
121 PPRLVVYELR LTNFKSYAGT QIVGPFHPSF SSIVGPNGSG KSNVIDALLF VFGFRASKLR 180
181 QSKASALIHK SATHPSLDSC DVEITFKEVN SDFTYVDGSE LTVRRTAYKN NTSKYFVNGV 240
241 ESSFSAVSNL LKEKGIDLNH KRFLILQGEV ESIAQMKPRA ISEGDDGLLE YLEDIIGTSK 300
301 YKPIIEENMQ ELSNSDDICA EKESRLKLVL SEKAKLEDSK NSVLSFLKDE NELFMKQNQL 360
361 YRTILYETRN KKTLVQNLLN SLEGKLQAHL EKFEQTERDI SEKNEEVKSL REKAAKVKND 420
421 CTSEKKTRQS YEQQTVKIEE QLKFLLNKEK KLKKSIEALS FEKSEAENSL SSHDIDSQKL 480
481 NSEIADLSLR LQQEELSLDD IRKSLQGKTE GISNAIEEKQ KAMAPALEKI NQLTSEKQIL 540
541 QVELDMLLNK ENDLINDVES SQSSLDKLRN DAEENRNILS SKLKVLSDLK GEKKDVSKNI 600
601 ERKKETVHNT YRNLMSNRTK LEEMKASLSS SRSRGNVLES LQRLHESDNL NGFFGRLGDL 660
661 ATIDEAYDVA ISTACPALNH IVVDNIETGQ KCVAFLRSNN LGRASFIILK ELAQKNLARI 720
721 QTPENVPRLF DLLRFNDQKF APAFYNVLQN TLVAKNLEQA NRIAYGKTRW RVVTLSGQLI 780
781 DKSGTMTGGG TRVKKGGMSS AITSDVSPAS VETCDKQVQL EDTRYRQHLS ELESLNQRFT 840
841 EISERIPSAE LEISKLQLDV SACDRLVAGE ERRILQLKSD LKSIRNNNER KRNLQNKISN 900
901 MDKEVEAINI NNEGLVTEIK TLQDKIMEIG GIRYRIQKSK VDDLHEQLKF VKDKLNKMSF 960
961 KKKKNEQRSQ SFQVELSNLT SEYDTTTESI ATLKTELQSL NKYVDEHKSR LREFENALWD1020
1021 INSSIDELVK FIEFESKQMN SVKAERIELE NQIQEQRTAL SEVGNNENKY LKLMSNLKLH1080
1081 NLTEFCDQTT MDSTFPEYSE DELSSVDKSE LVSNISVLKK KTEDREVDIN VLSEYRRCNK1140
1141 EAEKRDSDYQ SELQKRTDLK KVVTDLQSQR LDEFMYGFGI ISMKLKEMYQ IITMGGNAEL1200
1201 ELVDSLDPFS EGVLFSVMPP KKSWKNISNL SGGEKTLSSL ALVFALHNYK PTPLYVMDEI1260
1261 DAALDFKNVS IVANYIKERT KNAQFIVISL RSNMFELSSR LVGIYKTANM TKSVTINNKE1320
1321 ILTD |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Unpublished Data |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
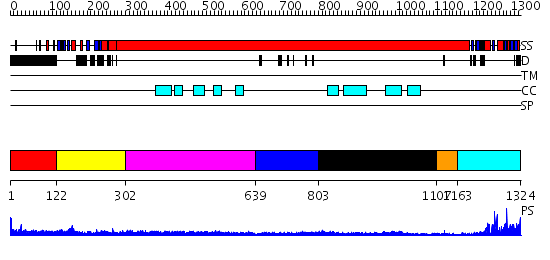
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..121] | 3.689995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [122..301] | 16.522879 | Sc Smc1hd:Scc1-C complex, ATPgS | |
| 3 | View Details | [302..638] | 11.09691 | Heavy meromyosin subfragment | |
| 4 | View Details | [639..802] | 4.1 | Smc hinge domain | |
| 5 | View Details | [803..1106] | 2.39 | Tropomyosin | |
| 6 | View Details | [1107..1162] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1163..1324] | 17.39794 | Structural biochemistry of ATP-driven dimerization and DNA stimulated activation of SMC ATPases. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)