













| General Information: |
|
| Name(s) found: |
rad22 /
SPAC30D11.10
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 469 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC |
| Biological Process: |
double-strand break repair via homologous recombination
[IGI protein homooligomerization [IDA gene conversion at mating-type locus, DNA repair synthesis [IMP meiotic DNA repair synthesis [IMP DNA recombinase assembly [NAS] mating type switching [IMP |
| Molecular Function: |
protein binding
[IPI damaged DNA binding [TAS single-stranded DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFEQKQHVA SEDQGHFNTA YSHEEFNFLQ SSLTRKLGPE YVSRRSGPGG FSVSYIESWK 60
61 AIELANEIFG FNGWSSSIRS INVDFMDENK ENGRISLGLS VIVRVTIKDG AYHEDIGYGS 120
121 IDNCRGKASA FEKCKKEGTT DALKRALRNF GNSLGNCMYD KYYLREVGKM KPPTYHFDSG 180
181 DLFRKTDPAA RESFIKKQKT LNSTRTVNNQ PLVNKGEQLA PRRAAELNDE QTREIEMYAD 240
241 EELDNIFVED DIIAHLAVAE DTAHPAANNH HSEKAGTQIN NKDKGSHNSA KPVQRSHTYP 300
301 VAVPQNTSDS VGNAVTDTSP KTLFDPLKPN TGTPSPKFIS ARAAAAAEGV VSAPFTNNFN 360
361 PRLDSPSIRK TSIIDHSKSL PVQRASVLPI IKQSSQTSPV SNNSMIRDSE SIINERKENI 420
421 GLIGVKRSLH DSTTSHNKSD LMRTNSDPQS AMRSRENYDA TVDKKAKKG |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
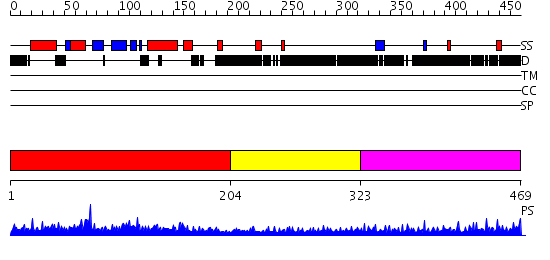
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..203] | 80.0 | The homologous-pairing domain of Rad52 recombinase | |
| 2 | View Details | [204..322] | 1.173997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [323..469] | 7.173992 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)