













| General Information: |
|
| Name(s) found: |
mis6 /
SPAC1687.20c
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 672 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC inner kinetochore of condensed chromosome [IDA spindle pole body [IDA condensed chromosome, centromeric region [IDA cytosol [IDA Mis6-Sim4 complex [TAS |
| Biological Process: |
sister chromatid segregation
[IMP mitotic spindle organization [IMP protein localization to kinetochore [IGI kinetochore assembly [TAS CenH3-containing nucleosome assembly at centromere [TAS cellular protein localization [IGI |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MESFENKGFL DIEEGIQWIK KNSENLSSSK KTILARLQQF HKLCSEVGLN QHSISSLLDV 60
61 ILSKKTHFDK NHVQLLIKCL YPNELISQTI AIRIISSLDP HGLRCSYAIQ AKLLNWLIHV 120
121 YEFLDGNNLL CRYYGVLFHF LDFLTLRPYI SNLLVLLTKH YHVKSFRIHQ LLALYQKPGN 180
181 TADPYLLALI LTYKQHFPDV IVGSYTYRKH GSVRLDSEWI AATKAILNRQ SEDVPLETWS 240
241 SEKRKRQSSL IPDLITMKNT SSSYSLEELT SVQQMGLVYE KIVFPSRIAA VLKSKLFLIF 300
301 LFLKNKNVYY SRLDEWLHIT LNYGLALRSG SNNQEEEVLH LLYKYLLFSP KFPKSLLQYV 360
361 ITFFSKPNIT EENYNLLTLL VTHIPITTDS SYFNSLLKEF EQFILQKNAE FCSKHLNILW 420
421 LWLFRMLNLR IASMGNNHTL LEKCLLITNH ATFLVSHFSW DVSLAYQLSR LFQLYYKILT 480
481 KIRKQIEPNI PPKELIYVLF FQPSAFYINS MVGLLLLTKN YQERLMDSRI DAISKFTHSY 540
541 LKSLSEIILL KEKRAILSFL QLWEPFKSDY SQFLPIATRI ANDHPYAQRV FSLTCAPQFF 600
601 SYINGYQIYL QQTNPATGSI PLKPIQEETF GAFQSNLHLS DSWEDFQKNF IIYLKKKGYL 660
661 AISDFLLSTL NR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Liu X, et al. (2005) | |
| View Run | No Comments | Liu X, et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
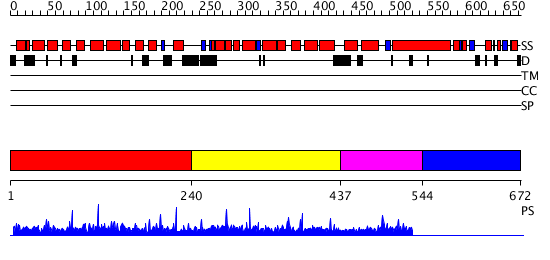
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..239] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [240..436] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [437..543] | N/A | Confident ab initio structure predictions are available. | |
| 4 | View Details | [544..672] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)