













| General Information: |
|
| Name(s) found: |
mid2 /
SPAPYUG7.03c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 704 amino acids |
Gene Ontology: |
|
| Cellular Component: |
medial cortex
[IDA cell division site [IDA cytoplasm [IDA cytosol [IDA septin ring [IDA |
| Biological Process: |
septin ring disassembly
[IMP cytokinesis [IMP cytokinetic cell separation [IMP septin ring organization [IMP |
| Molecular Function: |
GTP binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTASQQDQHA KMYLADIHRA LRIPSPIPST DYECSDYAST IASISRESTM RNFNRSNISS 60
61 TAPSFAESED AEDGDSFPYD QTLSNSSSFD DHQSLLPFST EVRRTPTYSV MNETDSSSTS 120
121 VEDVNKENIL SLNDSCLIKL SDDEASNKSS RSSTPRNSIK SNSSNQGHGD IPIPKKNPAR 180
181 SVCNSKLFNE DTLPAEFEEV SISPPVKLEL PTHSHNSSDT SFTNSIVSSV SDMVGLGEGI 240
241 NSIASFGFSE DSSSFQDIKT PPRLSFADEN RENCRTDIYR SDSIHEYEEP LTSSITSLDS 300
301 PHVLDENAPI PLLPKVVSLP DPRFTNVLSA FDALTRTYLL RQNSKVVHAT SQKQEMQTSR 360
361 RVVNSCYMPE SLSRNLSSSL QQTGGSGRLF VRLMEIRNLT IPLASGMTTR FTYTISGKHI 420
421 QVPWNALHST TKIENEYTFD ESISSSIVCT LRAAYDPPKV RTRSTLGKVF STNKRKSMTT 480
481 DPVSEALHGF VSEDGTFGEV TINTDSVSRT ALGRCQSMVL PIMNKWTVDP AAKDVKPLPR 540
541 KVGELEIHVF FLPALPVSLK ELPASIESAM YDLKLAEWDR TLLCDGYLCQ QGGDCPYWRR 600
601 RYFQLIGSKL VAFQQFSKVR RATIDLSEAT HIVDDNHYSD EEELEGYLYF ESGFRIIFSN 660
661 GDYIDFYAET VGEKDEWMST LRQHLGQCSM VHKNWTKSFL SLSF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
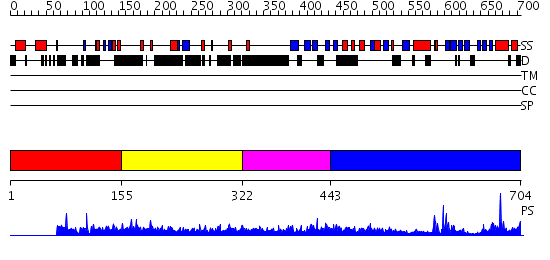
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..154] | 1.005943 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [155..321] | 2.046993 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [322..442] | 32.69897 | CYTOHESIN-1/B2-1 SEC7 DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | |
| 4 | View Details | [443..704] | 8.69897 | No description for 2q13A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)