













| General Information: |
|
| Name(s) found: |
Hsf-PA /
FBpp0085961
[FlyBase]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 691 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][ISS][NAS]
polytene chromosome [IDA] |
| Biological Process: |
response to heat
[IDA][NAS][TAS]
regulation of transcription, DNA-dependent [IEA] positive regulation of gene-specific transcription from RNA polymerase II promoter [IDA] |
| Molecular Function: |
DNA binding
[IDA][IEP][NAS][TAS]
transcription factor activity [IEA] specific RNA polymerase II transcription factor activity [IDA][ISS] sequence-specific DNA binding [IEA] transcription activator activity [IDA][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRSRSSAKA VQFKHESEEE EEDEEEQLPS RRMHSYGDAA AIGSGVPAFL AKLWRLVDDA 60
61 DTNRLICWTK DGQSFVIQNQ AQFAKELLPL NYKHNNMASF IRQLNMYGFH KITSIDNGGL 120
121 RFDRDEIEFS HPFFKRNSPF LLDQIKRKIS NNKNGDDKGV LKPEAMSKIL TDVKVMRGRQ 180
181 DNLDSRFSAM KQENEVLWRE IASLRQKHAK QQQIVNKLIQ FLITIVQPSR NMSGVKRHVQ 240
241 LMINNTPEID RARTTSETES ESGGGPVIHE LREELLDEVM NPSPAGYTAA SHYDQESVSP 300
301 PAVERPRSNM SISSHNVDYS NQSVEDLLLQ GNGTAGGNIL VGGAASPMAQ SVSQSPAQHD 360
361 VYTVTEAPDS HVQEVPNSPP YYEEQNVLTT PMVREQEQQK RQQLKENNKL RRQAGDVILD 420
421 AGDILVDSSS PKAQRTSIQH STQPDVMVQP MIIKSEPENS SGLMDLMTPA NDLYSVNFIS 480
481 EDMPTDIFED ALLPDGVEEA AKLDQQQKFG QSTVSSGKFA SNFDVPTNST LLDANQASTS 540
541 KAAAKAQASE EEGMAVAKYS GAENGNNRDT NNSQLLRMAS VDELHGHLES MQDELETLKD 600
601 LLRGDGVAID QNMLMGLFND SDLMDNYGLS FPNDSISSEK KAPSGSELIS YQPMYDLSDI 660
661 LDTDDGNNDQ EASRRQMQTQ SSVLNTPRHE L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
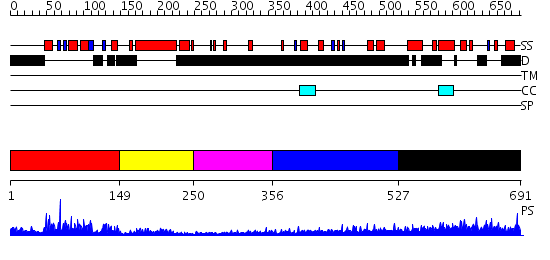
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..148] | 39.69897 | SOLUTION STRUCTURE OF THE DNA-BINDING DOMAIN OF DROSOPHILA HEAT SHOCK TRANSCRIPTION FACTOR | |
| 2 | View Details | [149..249] | 10.102373 | No description for PF00447.8 was found. No confident structure predictions are available. | |
| 3 | View Details | [250..355] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [356..526] | 9.239996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [527..691] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)