













| General Information: |
|
| Name(s) found: |
NMD2_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 1049 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSREEQIKKL NQYLDNRELA FRAKDGDKNI FHTESQLDSS LKKNTAFMKR CKSSLTSENY 60
61 DSFIKEIKTL SLKKFIPEIT AAIVEGMMKC KATKDILSSV KIVWALNLRF STAFTGPMLA 120
121 NLYCALYPNP GYSLCHESYF ELKQNENEVS EKDRSSHLLK VRPLLRFLIE FWLNGVVGTP 180
181 EDFVSYLPST DSNDKKFRKP WFEEQNLKKP LVVLLFNDLM DTRFGFLLLP VLTSLVRTFS 240
241 CELFTTEDFE DKETLELVNR LNPVVWRTYL RKSLNSYVDK LEVYCQKRKS LFEELNKQYQ 300
301 EQSIIRADPN NEKFQRLANF SKSIESEFSS YASLSEVLNR KASEDLLELN FMEKASSGTN 360
361 SVFNASGERS ESANVETAQV WDDREQYFFY EVFPNFNEGS IAEMKSSIYE SSQEGIRSSS 420
421 ENNKKEDDLK DSTGDLNTTQ VSSRVDNFLL KLPSMVSLEL TNEMALEFYD LNTKASRNRL 480
481 IKALCTIPRT SSFLVPYYVR LARILSQLSS EFSTSLVDHA RHSFKRMIHR KAKHEYDTRL 540
541 LIVRYISELT KFQLMPFHMV FECYKLCINE FTPFDLEVLA LLLESCGRFL LRYPETKLQM 600
601 QSFLEAIQKK KLASALASQD QLVLENALHF VNPPKRGIIV SKKKSLKEEF LYDLIQIRLK 660
661 DDNVFPTLLL LRKFDWKDDY QILYNTIMEV WNIKYNSLNA LARLLSALYK FHPEFCIHVI 720
721 DDTLESLFSA VNNSDHVEKQ KRLAQARFIS ELCVIHMLDV RAITNFLFHL LPLEKFESFL 780
781 TMKASTLTNI NNDMFRLRLI VVVLQTCGPS IIRSKTKKTM LTYLLAYQCY FLIQPEMPLD 840
841 MLYEFEDVIG YVRPSMKVYM HYEEARNALT ERLQAISDDW EEDDTRPVFQ GANDGDISSN 900
901 EESVYLPEDI SDESETDEES SGLEESDLLD SEDEDIDNEM QLSRELDEEF ERLTNESLLT 960
961 RMHEKNPGFD VPLPLRASSL GSPYVTRNEE SASESSHVMF TLLTKRGNKQ RSQYLEIPSH1020
1021 SSLVRSTKNQ QTEEIMERKR VKEMVLNFE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
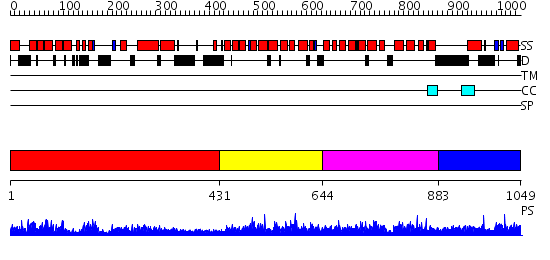
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..430] | 1.18 | Ribonuclease domain of colicin E3; Colicin E3 translocation domain; Colicin E3 receptor domain | |
| 2 | View Details | [431..643] | 48.09691 | Eukaryotic initiation factor eIF4G | |
| 3 | View Details | [644..882] | 76.522879 | The structural basis of the interaction between nonsense mediated decay factors UPF2 and UPF3 | |
| 4 | View Details | [883..1049] | 3.025983 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)