













| General Information: |
|
| Name(s) found: |
gi|12697967
[NCBI NR]
|
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 1090 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 LCKVSAGNEN ACLTTKHLTA LGKLEPKLVP LVIAFRYWAK LCSIDRPEEG GLPPYVFALM 60
61 AIFFLQQRKE PLLPVYLGSW IEGFSLSKLG NFNLQDIEKD VVIWEHTDSA AGDTGITKEE 120
121 APRETPIKRG QVSLILDVKH QPSVPVGQLW VELLRFYALE FNLADLVISI RVKELVSREL 180
181 KDWPKKRIAI EDPYSVKRNV ARTLNSQPVF EYILHCLRTT YKYFALPHKI TKSSLLKPLN 240
241 AITCISEHSK EVINHHPDVQ TKDDKLKNSV LAQGPGATSS AANTCKVQPL TLKETAESFG 300
301 SPPKEEMGNE HISVHPENSD CIQADVNSDD YKGDKVYHPE TGRKNEKEKV GRKGKHLLTV 360
361 DQKRGEHVVC GSTRNNESES TLDLEGFQNP TAKECEGLAT LDNKADLDGE STEGTEELED 420
421 SLNHFTHSVQ GQTSEMIPSD EEEEDDEEEE EEEEPRLTIN QREDEDGMAN EDELDNTYTG 480
481 SGDEDALSEE DDELGEAAKY EDVKECGKHV ERALLVELNK ISLKEENVCE EKNSPVDQSD 540
541 FFYEFSKLIF TKGKSPTVVC SLCKREGHLK KDCPEDFKRI QLEPLPPLTP KFLNILDQVC 600
601 IQCYKDFSPT IIEDQAREHI RQNLESFIRQ DFPGTKLSLF GSSKNGFGFK QSDLDVCMTI 660
661 NGLETAEGLD CVRTIEELAR VLRKHSGLRN ILPITTAKVP IVKFFHLRSG LEVDISLYNT 720
721 LALHNTRLLS AYSAIDPRVK YLCYTMKVFT KMCDIGDASR GSLSSYAYTL MVLYFLQQRN 780
781 PPVIPVLQEI YKGEKKPEIF VDGWNIYFFD QIDELPTYWS ECGKNTESVG QLWLGLLRFY 840
841 TEEFDFKEHV ISIRRKSLLT TFKKQWTSKY IVIEDPFDLN HNLGAGLSRK MTNFIMKAFI 900
901 NGRRVFGIPV KGFPKDYPSK MEYFFDPDVL TEGELAPNDR CCRICGKIGH FMKDCPMRRK 960
961 VRRRRDQEDA LNQRYPENKE KRSKEDKEIH NKYTEREVST KEDKPIQCTP QKAKPMRAAA1020
1021 DLGREKILRP PVEKWKRQDD KDLREKRCFI CGREGHIKKE CPQFKGSSGS LSSKYMTQGK1080
1081 ASAKRTQQES |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
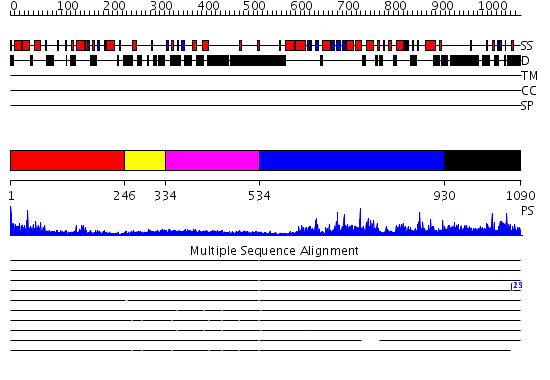
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..245] | 5.26 | Structural Basis for UTP Specificity of RNA Editing TUTases From Trypanosoma Brucei | |
| 2 | View Details | [246..333] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [334..533] | 1.279997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [534..929] | 64.522879 | Poly(A) polymerase, middle domain; Poly(A) polymerase N-terminal, catalytic domain; Poly(A) polymerase, C-terminal domain | |
| 5 | View Details | [930..1090] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)