













| General Information: |
|
| Name(s) found: |
RANBP9
[HGNC (HUGO)]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 729 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGQPPPPPP QQQQQQQQLS PPPPAALAPV SGVVLPAPPA VSAGSSPAGS PGGGAGGEGL 60
61 GAAAAALLLH PPPPPPPATA APPPPPPPPP PPASAAAPAS GPPAPPGLAA GPGPAGGAPT 120
121 PALVAGSSAA APFPHGDSAL NEQEKELQRR LKRLYPAVDE QETPLPRSWS PKDKFSYIGL 180
181 SQNNLRVHYK GHGKTPKDAA SVRATHPIPA ACGIYYFEVK IVSKGRDGYM GIGLSAQGVN 240
241 MNRLPGWDKH SYGYHGDDGH SFCSSGTGQP YGPTFTTGDV IGCCVNLINN TCFYTKNGHS 300
301 LGIAFTDLPP NLYPTVGLQT PGEVVDANFG QHPFVFDIED YMREWRTKIQ AQIDRFPIGD 360
361 REGEWQTMIQ KMVSSYLVHH GYCATAEAFA RSTDQTVLEE LASIKNRQRI QKLVLAGRMG 420
421 EAIETTQQLY PSLLERNPNL LFTLKVRQFI EMVNGTDSEV RCLGGRSPKS QDSYPVSPRP 480
481 FSSPSMSPSH GMNIHNLASG KGSTAHFSGF ESCSNGVISN KAHQSYCHSN KHQSSNLNVP 540
541 ELNSINMSRS QQVNNFTSND VDMETDHYSN GVGETSSNGF LNGSSKHDHE MEDCDTEMEV 600
601 DSSQLRRQLC GGSQAAIERM IHFGRELQAM SEQLRRDCGK NTANKKMLKD AFSLLAYSDP 660
661 WNSPVGNQLD PIQREPVCSA LNSAILETHN LPKQPPLALA MGQATQCLGL MARSGIGSCA 720
721 FATVEDYLH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
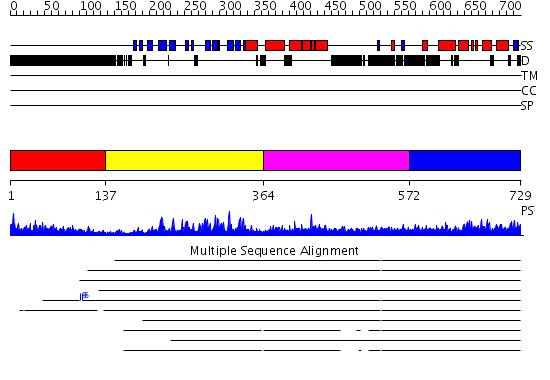
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..136] | 6.69897 | No description for 1y0fB was found. | |
| 2 | View Details | [137..363] | 26.09691 | No description for 2fnjA was found. | |
| 3 | View Details | [364..571] | 3.126977 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [572..729] | 3.044961 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 |
|
||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.55 |
Source: Reynolds et al. (2008)