













| General Information: |
|
| Name(s) found: |
SGT2 /
YOR007C
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 346 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
response to heat
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSASKEEIAA LIVNYFSSIV EKKEISEDGA DSLNVAMDCI SEAFGFEREA VSGILGKSEF 60
61 KGQHLADILN SASRVPESNK KDDAENVEIN IPEDDAETKA KAEDLKMQGN KAMANKDYEL 120
121 AINKYTEAIK VLPTNAIYYA NRAAAHSSLK EYDQAVKDAE SAISIDPSYF RGYSRLGFAK 180
181 YAQGKPEEAL EAYKKVLDIE GDNATEAMKR DYESAKKKVE QSLNLEKTVP EQSRDADVDA 240
241 SQGASAGGLP DLGSLLGGGL GGLMNNPQLM QAAQKMMSNP GAMQNIQKMM QDPSIRQMAE 300
301 GFASGGGTPN LSDLMNNPAL RNMAGNLFGG AGAQSTDETP DNENKQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
SGT2 YBR137W |
| View Details | Krogan NJ, et al. (2006) |
|
GET4 MDY2 NIP100 SGT2 YBR137W YDR026C |
| View Details | Ho Y, et al. (2002) |
|
CBK1 ECM10 GAL7 PRB1 SGT2 SSD1 UBP15 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample zds1 from august 2005 | McCusker D, et al (2007) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
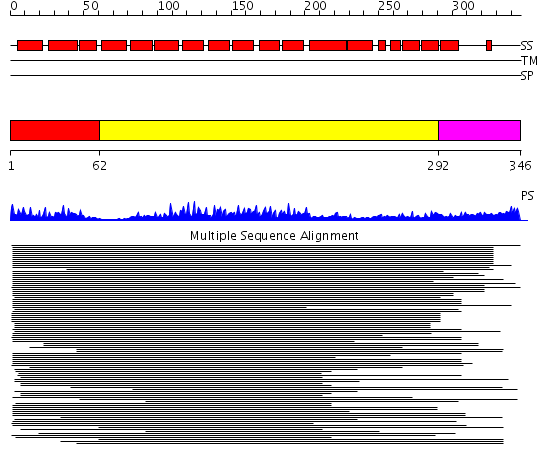
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..61] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [62..291] | 173.9691 | Protein phosphatase 5 | |
| 3 | View Details | [292..346] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)