













| General Information: |
|
| Name(s) found: |
MET1 /
YKR069W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 593 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IC |
| Biological Process: |
methionine biosynthetic process
[IMP siroheme biosynthetic process [IGI sulfate assimilation [NAS |
| Molecular Function: |
uroporphyrin-III C-methyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVRDLVTLPS SLPLITAGFA TDQVHLLIGT GSTDSVSVCK NRIHSILNAG GNPIVVNPSS 60
61 PSHTKQLQLE FGKFAKFEIV EREFRLSDLT TLGRVLVCKV VDRVFVDLPI TQSRLCEEIF 120
121 WQCQKLRIPI NTFHKPEFST FNMIPTWVDP KGSGLQISVT TNGNGYILAN RIKRDIISHL 180
181 PPNISEVVIN MGYLKDRIIN EDHKALLEEK YYQTDMSLPG FGYGLDEDGW ESHKFNKLIR 240
241 EFEMTSREQR LKRTRWLSQI MEYYPMNKLS DIKLEDFETS SSPNKKTKQE TVTEGVVPPT 300
301 DENIENGTKQ LQLSEVKKEE GPKKLGKISL VGSGPGSVSM LTIGALQEIK SADIILADKL 360
361 VPQAILDLIP PKTETFIAKK FPGNAERAQQ ELLAKGLESL DNGLKVVRLK QGDPYIFGRG 420
421 GEEFNFFKDH GYIPVVLPGI SSSLACTVLA QIPATQRDIA DQVLICTGTG RKGALPIIPE 480
481 FVESRTTVFL MALHRANVLI TGLLKHGWDG DVPAAIVERG SCPDQRVTRT LLKWVPEVVE 540
541 EIGSRPPGVL VVGKAVNALV EKDLINFDES RKFVIDEGFR EFEVDVDSLF KLY |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
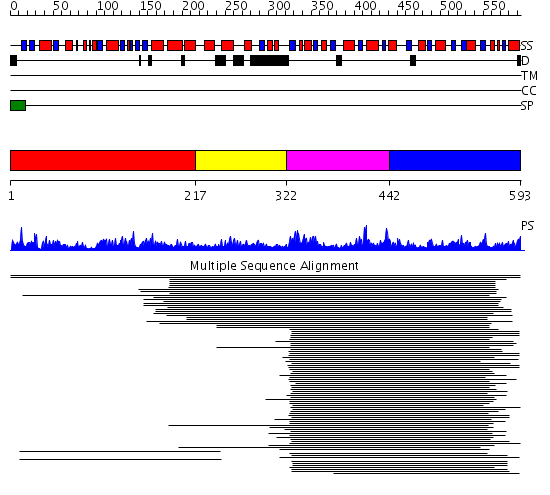
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..216] | 34.522879 | Bifunctional dehydrogenase/ferrochelatase Met8p, N-terminal domain; Bifunctional dehydrogenase/ferrochelatase Met8p, dimerisation and C-terminal domains | |
| 2 | View Details | [217..321] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [322..441] | 213.70927 | Cobalt precorrin-4 methyltransferase CbiF | |
| 4 | View Details | [442..593] | 213.70927 | Cobalt precorrin-4 methyltransferase CbiF |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|