













| General Information: |
|
| Name(s) found: |
YKL222C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 705 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ribosome
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKNTELSQKK KLDRQSRRKP AKSCHFCRVR KLKCDRVRPF CGSCSSRNRK QCEYKENTSA 60
61 MEDQLRKKYR RCSKLEMARR IEELESQLTK QSQPNIHEGQ NPLSNMRYLS SKHNRHILYG 120
121 PTSYRAILAT QTDTFAKYRE EIWQVLKLSR NNWKREHHYS TLSEISSIET APPHSGSPSV 180
181 IEYLCESLPN YEVLCEYLTD FFASDFYDSY QIVHKEKVLR DLQDCFVKGP RSHKTGQHTI 240
241 ISLNLDSKKN YYKVGVMTAI MCLASHPKEV PEAIEVFHKV LTSFVSAKVF YTERVQFLFL 300
301 RYLYINVAGL DGGDQSHCIF IHGLTIDTAI HMGLNEDLRR LYLSKNHPIE EIPYLERLWL 360
361 WILFTDVKIS LSTGIPVRIN DDFVNKVRLE NYSSSGDILL YKTTLRLRNI MKQIHAREKP 420
421 PDIPLIIEDL KKFTIKMFKP LDFYLNASNL NGNEFTELQL WHATLHMIGS LSNLYTLTHQ 480
481 DFDARIFNFS VLAPLNSLHL CFNVLETYFE LDNSKLSSKS LCLSKKWPHL NNALFLIYVN 540
541 AFRALIQIYT IFLQYMENKD IQLFIQRNSS ALTYSICPGD FEGPHNKCIS LKIAFKEMEN 600
601 IFDHIHQEKL KPLTQIWQNS YYFSIIISME KIGRRAFNKG MKNIDEGPET ENDATENSLT 660
661 TILNDLEGPL EDFSENFIDD ILGSPSAFFD TAISGWSNFE DFFSR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
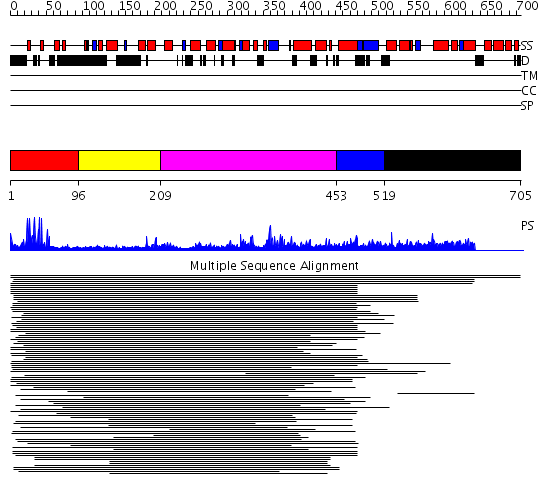
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..95] | 54.94696 | Hap1 (Cyp1); HAP1 | |
| 2 | View Details | [96..208] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [209..452] | 3.100987 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [453..518] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [519..705] | 1.010967 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)