













| General Information: |
|
| Name(s) found: |
FYV10 /
YIL097W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 516 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
negative regulation of gluconeogenesis
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEKSIFNEP DVDFHLKLNQ QLFHIPYELL SKRIKHTQAV INKETKSLHE HTAALNQIFE 60
61 HNDVEHDELA LAKITEMIRK VDHIERFLNT QIKSYCQILN RIKKRLEFFH ELKDIKSQNS 120
121 GTSHNGNNEG TRTKLIQWYQ SYTNILIGDY LTRNNPIKYN SETKDHWNSG VVFLKQSQLD 180
181 DLIDYDVLLE ANRISTSLLH ERNLLPLISW INENKKTLTK KSSILEFQAR LQEYIELLKV 240
241 DNYTDAIVCF QRFLLPFVKS NFTDLKLASG LLIFIKYCND QKPTSSTSSG FDTEEIKSQS 300
301 LPMKKDRIFQ HFFHKSLPRI TSKPAVNTTD YDKSSLINLQ SGDFERYLNL LDDQRWSVLN 360
361 DLFLSDFYSM YGISQNDPLL IYLSLGISSL KTRDCLHPSD DENGNQETET ATTAEKEVED 420
421 LQLFTLHSLK RKNCPVCSET FKPITQALPF AHHIQSQLFE NPILLPNGNV YDSKKLKKLA 480
481 KTLKKQNLIS LNPGQIMDPV DMKIFCESDS IKMYPT |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
FYV10 GID7 GID8 RMD5 VID24 VID28 VID30 YDL176W |
| View Details | Krogan NJ, et al. (2006) |
|
FYV10 GID7 GID8 RMD5 VID24 VID28 VID30 YDL176W |
| View Details | Ho Y, et al. (2002) |
|
CCT2 CCT3 CTF19 FYV10 GID7 GID8 HXT7 MOH1 RMD5 SHS1 TFP1 UME1 VID24 VID28 VID30 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
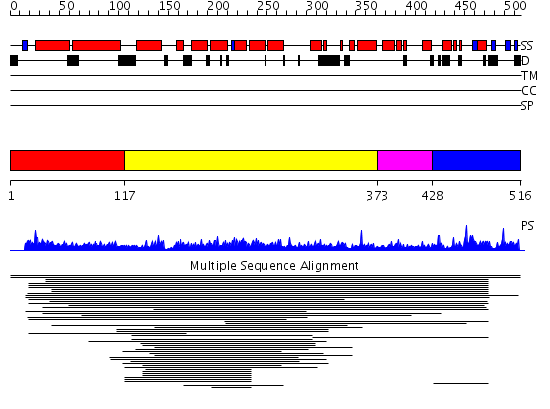
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..116] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [117..372] | 9.039979 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [373..427] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [428..516] | 1.016986 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)