













| General Information: |
|
| Name(s) found: |
RSC30 /
YHR056C
[SGD]
|
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 832 amino acids |
Gene Ontology: |
|
| Cellular Component: |
RSC complex
[IDA |
| Biological Process: |
ATP-dependent chromatin remodeling
[IDA regulation of transcription, DNA-dependent [IMP double-strand break repair via nonhomologous end joining [IDA |
| Molecular Function: |
DNA binding
[ISS DNA-dependent ATPase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVAVPSASGM STHGNGQGSN HFSQGNGVNQ KNVMIQTQYP IMQTSIEAFN FSFNPSVDTA 60
61 MQWTKAASYQ NNNTNNNTAP RQNSSTVSSN VHGNTIVRSD SPDVPSMDQI REYNTRLQLV 120
121 NAQSFDYTDN PYSFNVGINQ DSAVFDLMTS PFTQEEVLIK EIDFLKNKLL DLQSLQLKSL 180
181 KEKSNLNADN TTANKINKTG ENSKKGKVDG KRAGFDHQTS RTSQSSQKYF TALTITDVQS 240
241 LVQVKPLKDT PNYLFTKNFI IFRDHYLFKF YNILHDICHI NQFKVSPPNN KNHQQYMEVC 300
301 KVNFPPKAII IETLNSESLN NLNIEEFLPI FDKTLLLEFV HNSFPNGDTC PSFSTVDLPL 360
361 SQLTKLGELT VLLLLLNDSM TLFNKQAINN HVSALMNNLR LIRSQITLIN LEYYDQETIK 420
421 FIAITKFYES LYMHDDHKSS LDEDLSCLLS FQIKDFKLFH FLKKMYYSRH SLLGQSSFMV 480
481 PAAENLSPIP ASIDTNDIPL IANDLKLLET QAKLINILQG VPFYLPVNLT KIESLLETLT 540
541 MGVSNTVDLY FHDNEVRKEW KDTLNFINTI VYTNFFLFVQ NESSLSMAVQ HSSNNNKTSN 600
601 SERCAKDLMK IISNMHIFYS ITFNFIFPIK SIKSFSSGNN RFHSNGKEFL FANHFIEILQ 660
661 NFIAITFAIF QRCEVILYDE FYKNLSNEEI NVQLLLIHDK ILEILKKIEI IVSFLRDEMN 720
721 SNGSFKSIKG FNKVLNLIKY MLRFSKKKQN FARNSDNNNV TDYSQSAKNK NVLLKFPVSE 780
781 LNRIYLKFKE ISDFLMEREV VQRSIIIDKD LESDNLGITT ANFNDFYDAF YN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | 3912: TAP-tagged, strain background includes hpc2(delta) | Green EM, et al (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
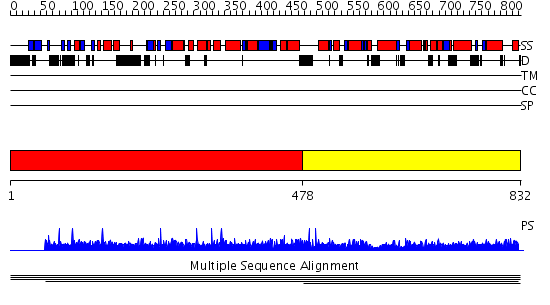
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..477] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [478..832] | 1.00386 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)