













| General Information: |
|
| Name(s) found: |
RIM101 /
YHL027W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 625 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC |
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter
[IDA ascospore formation [IMP fungal-type cell wall biogenesis [IGI response to pH [IMP meiosis [IMP |
| Molecular Function: |
specific transcriptional repressor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVPLEDLLNK ENGTAAPQHS RESIVENGTD VSNVTKKDGL PSPNLSKRSS DCSKRPRIRC 60
61 TTEAIGLNGQ EDERMSPGST SSSCLPYHST SHLNTPPYDL LGASAVSPTT SSSSDSSSSS 120
121 PLAQAHNPAG DDDDADNDGD SEDITLYCKW DNCGMIFNQP ELLYNHLCHD HVGRKSHKNL 180
181 QLNCHWGDCT TKTEKRDHIT SHLRVHVPLK PFGCSTCSKK FKRPQDLKKH LKIHLESGGI 240
241 LKRKRGPKWG SKRTSKKNKS CASDAVSSCS ASVPSAIAGS FKSHSTSPQI LPPLPVGISQ 300
301 HLPSQQQQRA ISLNQLCSDE LSQYKPVYSP QLSARLQTIL PPLYYNNGST VSQGANSRSM 360
361 NVYEDGCSNK TIANATQFFT KLSRNMTNNY ILQQSGGSTE SSSSSGRIPV AQTSYVQPPN 420
421 APSYQSVQGG SSISATANTA TYVPVRLAKY PTGPSLTEHL PPLHSNTAGG VFNRQSQYAM 480
481 PHYPSVRAAP SYSSSGCSIL PPLQSKIPML PSRRTMAGGT SLKPNWEFSL NQKSCTNDII 540
541 MSKLAIEEVD DESEIEDDFV EMLGIVNIIK DYLLCCVMED LDDEESEDKD EENAFLQESL 600
601 EKLSLQNQMG TNSVRILTKY PKILV |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
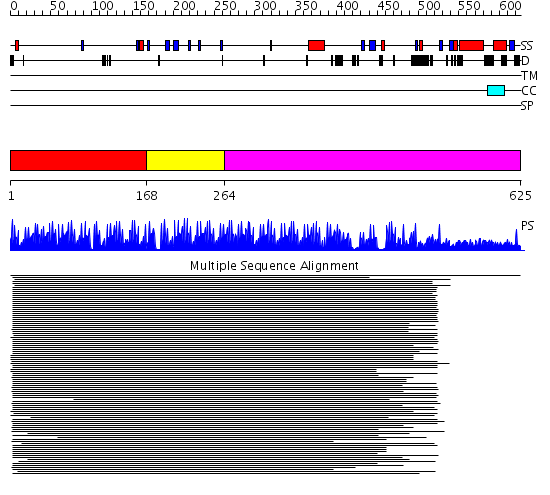
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..167] | 43.909996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [168..263] | 99.855515 | ZIF268 | |
| 3 | View Details | [264..625] | 7.16 | Monomethylamine methyltransferase MtmB |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)