













| General Information: |
|
| Name(s) found: |
YSW1 /
YBR148W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 609 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSLADTVEG SEAKRGRFSN NALTSDTGIL QKNSTLRNWF LKPTADLKNS CEDRVEDDVN 60
61 DVYLNDKNSQ KSVEERKLGR KVRSFFKQTN SNKDESVLED EDDALVWKKT SNKCAKKENS 120
121 HDIQKGSFTK KIRNSIFKSA NDVKEFRNEN NLLLPVELSS DDENESHFTD ANSHVMQSKS 180
181 PEKIPSKDQC LTKGAKNKGL KKEYEKSFEE YSDDSDDEFS PATPPENVLE GPYKFVFQTP 240
241 NTFTSQPNIT VENDFHKGGR HVIDYLNKKL ATMNIDIDLT SGGKQNVSWE EELDQLSDHV 300
301 IESITNHISK GRMHAQEKQD ELEKLKLENL NLSTLKQENL QHKQEINSLK DNLESISKKN 360
361 NDLILEMNKL KKKSTNNKTN EYISTDENEN EEITKSNMGP GILELNVNET SKKLQQSTFK 420
421 PSKYLPRETR NNENRLKHLE KRIFGLEKSL EKKKKQVRAD SVRLDLNRYT IDQFLTLLKS 480
481 LSEVLQFHNV YGNDLKENDD NIIKIETCCS ALNMKNCFED SSFRLQENSF KRQLGPLFAN 540
541 INFSLIDQLT MNFRFYERSA NFQKETIGGL RMMLQDKDNY IKTLMQHLKK KESTKLIKDS 600
601 KNGASTLTS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
NOG1 NOG2 NOP15 NUG1 YSW1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
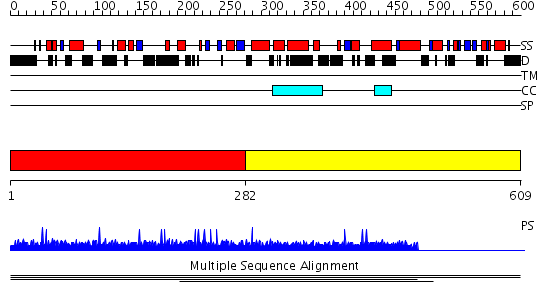
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..281] | 7.7 | Microbial transglutaminase | |
| 2 | View Details | [282..609] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)