













| General Information: |
|
| Name(s) found: |
NMD2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1089 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDDGRKKELH DLNTRAWNGE EVFPLKSKKL DSSIKRNTGF IKKLKKGFVK GSESSLLKDL 60
61 SEASLEKYLS EIIVTVTECL LNVLNKNDDV IAAVEIISGL HQRFNGRFTS PLLGAFLQAF 120
121 ENPSVDIESE RDELQRITRV KGNLRVFTEL YLVGVFRTLD DIESKDAIPN FLQKKTGRKD 180
181 PLLFSILREI LNYKFKLGFT TTIATAFIKK FAPLFRDDDN SWDDLIYDSK LKGALQSLFK 240
241 NFIDATFARA TELHKKVNKL QREHQKCQIR TGKLRDEYVE EYDKLLPIFI RFKTSAITLG 300
301 EFFKLEIPEL EGASNDDLKE TASPMITNQI LPPNQRLWEN EDTRKFYEIL PDISKTVEES 360
361 QSSKTEKDSN VNSKNINLFF TDLEMADCKD IIDDLSNRYW SSYLDNKATR NRILKFFMET 420
421 QDWSKLPVYS RFIATNSKYM PEIVSEFINY LDNGFRSQLH SNKINVKNII FFSEMIKFQL 480
481 IPSFMIFHKI RTLIMYMQVP NNVEILTVLL EHSGKFLLNK PEYKELMEKM VQLIKDKKND 540
541 RQLNMNMKSA LENIITLLYP PSVKSLNVTV KTITPEQQFY RILIRSELSS LDFKHIVKLV 600
601 RKAHWDDVAI QKVLFSLFSK PHKISYQNIP LLTKVLGGLY SYRRDFVIRC IDQVLENIER 660
661 GLEINDYGQN MHRISNVRYL TEIFNFEMIK SDVLLDTIYH IIRFGHINNQ PNPFYLNYSD 720
721 PPDNYFRIQL VTTILLNINR TPAAFTKKCK LLLRFFEYYT FIKEQPLPKE TEFRVSSTFK 780
781 KYENIFGNTK FERSENLVES ASRLESLLKS LNAIKSKDDR VKGSSASIHN GKESAVPIES 840
841 ITEDDEDEDD ENDDGVDLLG EDEDAEISTP NTESAPGKHQ AKQDESEDED DEDDDEDDDD 900
901 DDDDDDDDGE EGDEDDDEDD DDEDDDDEEE EDSDSDLEYG GDLDADRDIE MKRMYEEYER 960
961 KLKDEEERKA EEELERQFQK MMQESIDARK SEKVVASKIP VISKPVSVQK PLLLKKSEEP1020
1021 SSSKETYEEL SKPKKIAFTF LTKSGKKTQS RILQLPTDVK FVSDVLEEEE KLKTERNKIK1080
1081 KIVLKRSFD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
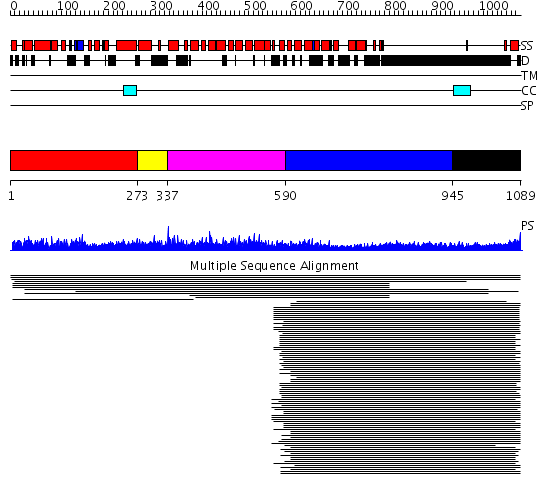
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..272] | 28.070581 | MIF4G domain No confident structure predictions are available. | |
| 2 | View Details | [273..336] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [337..589] | 13.8 | CBP80, 80KDa nuclear cap-binding protein | |
| 4 | View Details | [590..944] | 48.522879 | Heavy meromyosin subfragment | |
| 5 | View Details | [945..1089] | 17.221849 | Tropomyosin |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)