













| General Information: |
|
| Name(s) found: |
NCL1_YEAST
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 684 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MARRKNFKKG NKKTFGARDD SRAQKNWSEL VKENEKWEKY YKTLALFPED QWEEFKKTCQ 60
61 APLPLTFRIT GSRKHAGEVL NLFKERHLPN LTNVEFEGEK IKAPVELPWY PDHLAWQLDV 120
121 PKTVIRKNEQ FAKTQRFLVV ENAVGNISRQ EAVSMIPPIV LEVKPHHTVL DMCAAPGSKT 180
181 AQLIEALHKD TDEPSGFVVA NDADARRSHM LVHQLKRLNS ANLMVVNHDA QFFPRIRLHG 240
241 NSNNKNDVLK FDRILCDVPC SGDGTMRKNV NVWKDWNTQA GLGLHAVQLN ILNRGLHLLK 300
301 NNGRLVYSTC SLNPIENEAV VAEALRKWGD KIRLVNCDDK LPGLIRSKGV SKWPVYDRNL 360
361 TEKTKGDEGT LDSFFSPSEE EASKFNLQNC MRVYPHQQNT GGFFITVFEK VEDSTEAATE 420
421 KLSSETPALE SEGPQTKKIK VEEVQKKERL PRDANEEPFV FVDPQHEALK VCWDFYGIDN 480
481 IFDRNTCLVR NATGEPTRVV YTVCPALKDV IQANDDRLKI IYSGVKLFVS QRSDIECSWR 540
541 IQSESLPIMK HHMKSNRIVE ANLEMLKHLL IESFPNFDDI RSKNIDNDFV EKMTKLSSGC 600
601 AFIDVSRNDP AKENLFLPVW KGNKCINLMV CKEDTHELLY RIFGIDANAK ATPSAEEKEK 660
661 EKETTESPAE TTTGTSTEAP SAAN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
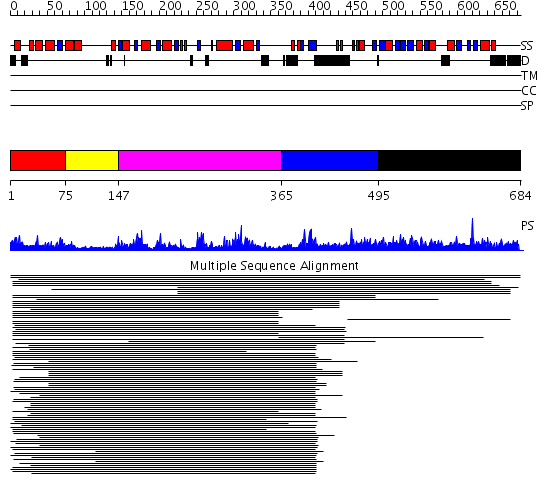
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..74] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [75..146] | 5.69897 | Fibrillarin homologue | |
| 3 | View Details | [147..364] | 164.10721 | No description for 1j4fA_ was found. | |
| 4 | View Details | [365..494] | 1.17399 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [495..684] | 1.01197 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)