













| General Information: |
|
| Name(s) found: |
gi|7494203
[NCBI NR]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Plasmodium falciparum |
| Length: | 955 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVTGGQGKLN QKNQKGDKKN LSYDNKINNN INISVVEKSE GNYKESYVFY KKDMCDENNN 60
61 ISNMEGIDQF HNCNNEQKKI DTTTITTYNN NINSSSNNNY NNDYDIDENN NFHCHDDDHK 120
121 HLDNYCVKDD YSHEDNDQCH VCNTVDMLNL EEERHFCNVC FSFLYYKKYC FYELLRIYKN 180
181 YSCLNEEEKN LLTESIYAKL YKMYLTTLNN YYFILNILLP QISTHIIIHL LTFTFHKPEE 240
241 DEHMKEEYVM NEIINKLTNQ ELNNIDKVYN YHNFNVRLLC KDDALIILNE IRSKMINSQN 300
301 EKKIVDMLNM KCGDVIVDDK NGSDKDSGNN DIHNKDIHNK YDDNMDSSDK SSCNKNSCDN 360
361 HCDNHCDNHS DNHCDNHSDN HCDNHCDNHC DNHCDNHCDN HCDNHCDNHC DNHCDNHCDN 420
421 HCDNHCDNHC DNHCDNHCDN HCDNHSDNHC DNHCDNHCDN HCDNHCDNHC DNHCDNHSDN 480
481 HCDNHCDNHS DNHCDNHCDN HSDNHCDNHC DNHCDNHCDN HCDNHCDNHC DNHCDNHCDN 540
541 HCDNHCDNHC DNHCDNHCDN HCDNHCDNHC DNNKMNRTHL MNSNTKGDDQ TCKNTNEYSD 600
601 TSSNHYCNNI EEDTYNVVNP GVDLDVHNII DSKNIKDYHN DIKENMCNNN KTLHADENMP 660
661 NDNICARENN SKYEHTPPGM RISQAYTPTL DDYNLIQNMS KVRSTLRQFV RDWSMEGQEE 720
721 RDKAYIPLIN SLDKYLPIHD NYIPKILCPG SGLGRLPYEI AKKGYRSQGN EFSYFMLLGS 780
781 NFILNYYNEK YSLSIHPYCL CTSNRRGRDD HLKIIQLPDV NTYNKIVLNT DFSMCAGELI 840
841 EVYYYDKEYF DGVLTCFFLD TAKNLFMYIR TFASILKPNS LWSNIGPLLY HYAEMPNEMS 900
901 IELAWDEIQI IISKWFTIKE IKWIDNYYTT NFDSMMQVQY HCVFFSAIRN DIPVD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
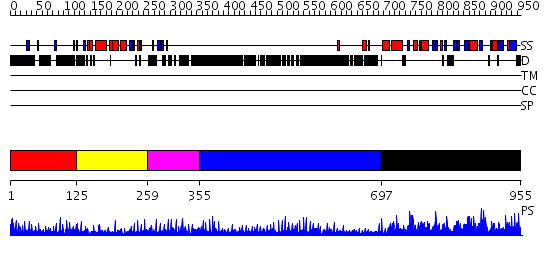
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..124] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [125..258] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [259..354] | 4.69897 | Hybrid domain of integrin beta; Integrin beta A domain; Integrin beta tail domain; Integrin beta EGF-like domains | |
| 4 | View Details | [355..696] | 32.522879 | Low density lipoprotein (LDL) receptor YWTD domain; Low density lipoprotein (LDL) receptor, different EGF domains; Ligand-binding domain of low-density lipoprotein receptor | |
| 5 | View Details | [697..955] | 1.89 | Crystal structure of thiopurine S-methyltransferase. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 |
|
|||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)