













| General Information: |
|
| Name(s) found: |
RH11_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 612 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisome
[IDA]
nucleolus [IDA] plasma membrane [IDA] |
| Biological Process: | NONE FOUND |
| Molecular Function: |
nucleic acid binding
[IEA]
ATP binding [IEA] ATP-dependent helicase activity [IEA][ISS] helicase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSASWADVAD SEKAVSQSKP PYVPPHLRNR PSEPVAAPLP QNDHAGYGGQ PAGSRWAPPS 60
61 SGGGGASGGG YRNDGGRTGY GYGAGGGGGG GGGWNNRSGG WDRREREVNP FGDDAELEPV 120
121 FTEQENTGIN FDAYEDIPVE TSGGDVPPPV NTFADIDLGD ALNLNIRRCK YVRPTPVQRH 180
181 AIPILLAERD LMACAQTGSG KTAAFCFPII SGIMKDQHVE RPRGSRAVYP FAVILSPTRE 240
241 LACQIHDEAK KFSYQTGVKV VVAYGGTPIH QQLRELERGC DILVATPGRL NDLLERARVS 300
301 MQMIRFLALD EADRMLDMGF EPQIRKIVEQ MDMPPRGVRQ TMLFSATFPS QIQRLAADFM 360
361 SNYIFLAVGR VGSSTDLITQ RVEFVQESDK RSHLMDLLHA QRETQDKQSL TLVFVETKRG 420
421 ADTLENWLCM NEFPATSIHG DRTQQEREVA LRSFKTGRTP ILVATDVAAR GLDIPHVAHV 480
481 VNFDLPNDID DYVHRIGRTG RAGKSGIATA FFNENNAQLA RSLAELMQEA NQEVPEWLTR 540
541 YASRASFGGG KKRSGGRFGG RDFRREGSYS RGGGGGGGGG GSDYYGGGGY GGGGYGGAPS 600
601 GGYGAGVTSA WD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
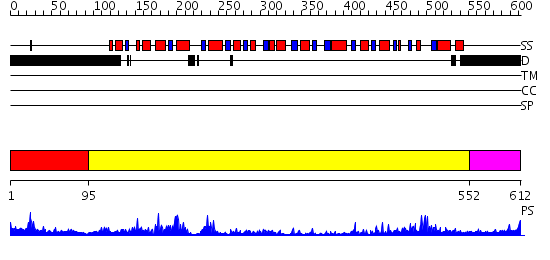
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..94] | 1.342996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [95..551] | 102.0 | No description for 2i4iA was found. | |
| 3 | View Details | [552..612] | 21.522879 | Nucleotide excision repair enzyme UvrB |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)