













| General Information: |
|
| Name(s) found: |
DIM1C_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 343 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IEA]
|
| Biological Process: |
response to cold
[IMP]
|
| Molecular Function: |
mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMNAVITSAT INCNSLSPSW TCGDNSPSKL LLGEISAALS RRRTVKVSCG KSSPDDYHST 60
61 LKSLNSRGRF PRKSLGQHYM LNSDINDQLA SAADVKEGDF VLEIGPGTGS LTNVLINLGA 120
121 TVLAIEKDPH MVDLVSERFA GSDKFKVLQE DFVKCHIRSH MLSILETRRL SHPDSALAKV 180
181 VSNLPFNIST DVVKLLLPMG DIFSKVVLLL QDEAALRLVE PALRTSEYRP INILINFYSE 240
241 PEYNFRVPRE NFFPQPKVDA AVVTFKLKHP RDYPDVSSTK NFFSLVNSAF NGKRKMLRKS 300
301 LQHISSSPDI EKALGVAGLP ATSRPEELTL DDFVKLHNVI ARE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
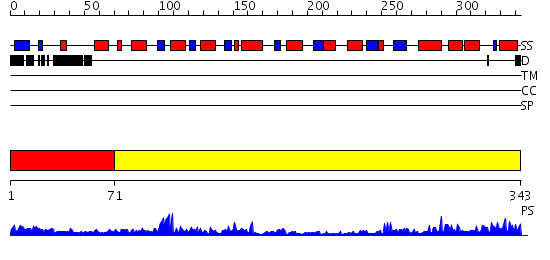
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..70] | 7.69897 | Crystal Structure Analysis of E.coli Protein (N5)-Glutamine Methyltransferase (HemK) | |
| 2 | View Details | [71..343] | 51.30103 | 2.1 Angstrom Crystal structure of KsgA: A Universally Conserved Adenosine Dimethyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)