













| General Information: |
|
| Name(s) found: |
KN12A_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1292 amino acids |
Gene Ontology: |
|
| Cellular Component: |
phragmoplast
[IDA]
|
| Biological Process: |
phragmoplast assembly
[IGI]
microgametogenesis [IMP] |
| Molecular Function: |
microtubule motor activity
[ISS]
plus-end-directed microtubule motor activity [IGI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKKHFTLPRN AILRDGGEPH SPNPSISKSK PPRKLRSAKE NAPPLDRNTS TPDHRSMRMK 60
61 NPLPPRPPPS NPLKRKLSAE TATESGFSDS GVKVIVRMKP LNKGEEGDMI VEKMSKDSLT 120
121 VSGQTFTFDS IANPESTQEQ MFQLVGAPLV ENCLSGFNSS VFAYGQTGSG KTYTMWGPAN 180
181 GLLEEHLCGD QRGLTPRVFE RLFARIKEEQ VKHAERQLNY QCRCSLLEIY NEQITDLLDP 240
241 SQKNLMIRED VKSGVYVENL TEEYVKNLTD VSQLLIKGLG NRRTGATSVN TESSRSHCVF 300
301 TCVVESRCKN VADGLSSFKT SRINLVDLAG SERQKSTGAA GERLKEAGNI NRSLSQLGNL 360
361 INILAEISQT GKPRHIPYRD SRLTFLLQES LGGNAKLAMV CAVSPSQSCR SETFSTLRFA 420
421 QRAKAIQNKA VVNEVMQDDV NFLRGVIHQL RDELQRMKND GNNPTNPNVA YSTAWNARRS 480
481 LNLLRSFGLG HPRSLPHEDN DGDIEMEIDE AAVERLCVQV GLQSSLASEG INHDMNRVKS 540
541 IHSSDGQSIE KRLPEDSDVA MEDACCHTEN HEPETVDNMR TETETGIREN QIKTHSQTLD 600
601 HESSFQPLSV KDALCSSLNK SEDVSSCPDL VPQDVTSANV LIADGVDDPE HLVNSASPSL 660
661 CIDPVGATPV LKSPTLSVSP TIRNSRKSLK TSELSTASQK DSEGENLVTE AADPSPATSK 720
721 KMNNCSSALS TQKSKVFPVR TERLASSLHK GIKLLESYCQ STAQRRSTYR FSFKAPDSEP 780
781 STSISKADAG VQTIPGADAI SEENTKEFLC CKCKCREQFD AQQMGDMPNL QLVPVDNSEV 840
841 AEKSKNQVPK AVEKVLAGSI RREMALEEFC TKQASEITQL NRLVQQYKHE RECNAIIGQT 900
901 REDKIIRLES LMDGVLSKED FLDEEFASLL HEHKLLKDMY QNHPEVLKTK IELERTQEEV 960
961 ENFKNFYGDM GEREVLLEEI QDLKLQLQCY IDPSLKSALK TCTLLKLSYQ APPVNAIPES1020
1021 QDESLEKTLE QERLCWTEAE TKWISLSEEL RTELEASKAL INKQKHELEI EKRCGEELKE1080
1081 AMQMAMEGHA RMLEQYADLE EKHMQLLARH RRIQDGIDDV KKAAARAGVR GAESRFINAL1140
1141 AAEISALKVE KEKERQYLRD ENKSLQTQLR DTAEAIQAAG ELLVRLKEAE EGLTVAQKRA1200
1201 MDAEYEAAEA YRQIDKLKKK HENEINTLNQ LVPQSHIHNE CSTKCDQAVE PSVNASSEQQ1260
1261 WRDEFEPLYK KETEFSNLAE PSWFSGYDRC NI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
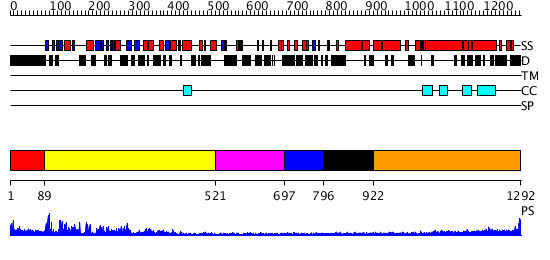
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..88] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [89..520] | 86.30103 | Kinesin | |
| 3 | View Details | [521..696] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [697..795] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [796..921] | N/A | Confident ab initio structure predictions are available. | |
| 6 | View Details | [922..1292] | 9.522879 | Heavy meromyosin subfragment |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)