













| General Information: |
|
| Name(s) found: |
RPN2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 691 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum membrane
[ISS]
mitochondrion [IDA] membrane [IDA] endoplasmic reticulum [IDA] plant-type cell wall [IDA] plasma membrane [IDA] |
| Biological Process: |
response to cold
[IEP]
protein amino acid terminal N-glycosylation [ISS] |
| Molecular Function: |
dolichyl-diphosphooligosaccharide-protein glycotransferase activity
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMAGGNVRFL VLILAVAICG AASVFQPISD SHRSAALDVF VPVDGSYKSL EEAYEALKTL 60
61 EILGIDKKSD LSSKTCENVV KVLQSSSSTL KDAFYALNVN GILKCKIGEA GPKDIVSQLQ 120
121 AGVKDAKLLL DFYYSVRGLV LAKEQFPGTH ISLGDAEAIF RSIKSLSQSD GRWRYSSNNP 180
181 ESSTFAAGLA YETLAGVISL APSEFDPSLI QSVKTGILKL SDSIQKYDDG TFYFDEKSVD 240
241 ASQGPISTTA SVIRGLTSFA ASESTGLNLP GDKIVGLAKF FLGVGIPGDA KDFFNQIDAL 300
301 ACLEDNKFSV PLILSLPSTV ISLTKKEPLK VKVSTVLGSK APALSVKLTQ ALSSKSVDSS 360
361 VINNQELKFD ADSATYFLDS FPKNFDIGKY TFVFKIVLDE SAHEKVYITE AQTKVPIAAT 420
421 GAISIENAEI AVLDSDIGSV ESQKKLDLTK DGAVSLSANH LQKLRLSFQL TTPLGNAFKP 480
481 HQAFFKLKHE SQVEHIFLVK TSGKKSELVL DFLGLVEKLY YLSGKYEIQL TIGDASMENS 540
541 LLSNIGHIEL DLPERPEKAT RPPLQSTEPY SRYGPKAEIS HIFRIPEKLP AKQLSLVFLG 600
601 VIVLPFIGFL IGLTRLGVNI KSFPSSTGSA ISALLFHCGI GAVLLLYVLF WLKLDLFTTL 660
661 KALSLLGVFL LFVGHRTLSQ LASASNKLKS A |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
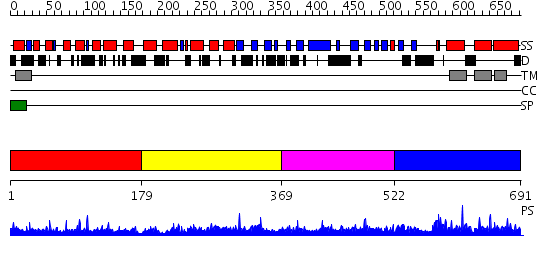
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..178] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [179..368] | 1.023965 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [369..521] | 1.039971 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [522..691] | 5.047968 | View MSA. No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|