













| General Information: |
|
| Name(s) found: |
DNJ49_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 354 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
protein folding
[IEA][ISS]
|
| Molecular Function: |
protein binding
[IPI]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDGNKDDASR CLRIAEDAIV SGDKERALKF INMAKRLNPS LSVDELVAAC DNLDSVSRNS 60
61 SVSEKLKTMD GDDDKLETGK MKYTEENVDL VRNIIRNNDY YAILGLEKNC SVDEIRKAYR 120
121 KLSLKVHPDK NKAPGSEEAF KKVSKAFTCL SDGNSRRQFD QVGIVDEFDH VQRRNRRPRR 180
181 RYNTRNDFFD DEFDPEEIFR TVFGQQREVF RASHAYRTRQ PRNQFREEEI NVAGPSCLTI 240
241 IQILPFFLLL LLAYLPFSEP DYSLHKNQSY QIPKTTQNTE ISFYVRSASA FDEKFPLSSS 300
301 ARANLEGNVI KEYKHFLFQS CRIELQKRRW NKKIPTPHCI ELQDRGFVDR HIPI |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
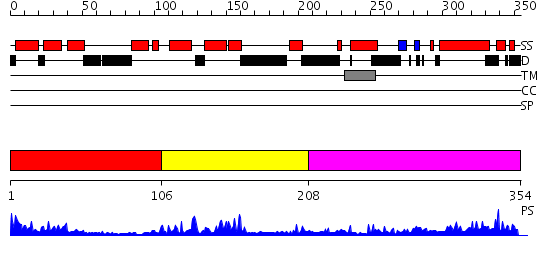
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..105] | 8.69897 | Designed protein cTPR3 | |
| 2 | View Details | [106..207] | 17.522879 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 3 | View Details | [208..354] | 7.30103 | The crystal structure of Hsp40 Ydj1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.27 |
Source: Reynolds et al. (2008)