













| General Information: |
|
| Name(s) found: |
gi|42564975
[NCBI NR]
gi|115646890 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 517 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IEA]
|
| Biological Process: |
response to heat
[IEA]
protein folding [IEA][ISS] |
| Molecular Function: |
ATP binding
[IEA]
heat shock protein binding [IEA] unfolded protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAMARCALI PSINPAHSFR HQFPQPNASF YLPPTLPIFS RVRRFGISGG YRRRVITMAA 60
61 GTDHYSTLNV NRNATLQEIK SSYRKLARKY HPDMNKNPGA EDKFKQISAA YEVLSDEEKR 120
121 SAYDRFGEAG LEGDFNGSQD TSPGVDPFDL YSAFFGGSDG FFGGMGESGG MGFDFMNKRS 180
181 LDLDIRYDLR LSFEEAVFGV KREIEVSYLE TCDGCGGTGA KSSNSIKQCS SCDGKGRVMN 240
241 SQRTPFGIMS QVSTCSKCGG EGKTITDKCR KCIGNGRLRA RKKMDVVVPP GVSDRATMRI 300
301 QGEGNMDKRS GRAGDLFIVL QVDEKRGIRR EGLNLYSNIN IDFTDAILGA TTKVETVEGS 360
361 MDLRIPPGTQ PGDTVKLPRK GVPDTDRPSI RGDHCFVVKI SIPKKLSERE RKLVEEFSSL 420
421 RRSSSSTGPT ETRQEEQSFG SEPRKEPSLW HKMKNFIRPE DSRTKFGTMS LNPSLPLRRM 480
481 KVSETSIAFS VLALCVITSA VALVQKKGNR LKQKKET |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
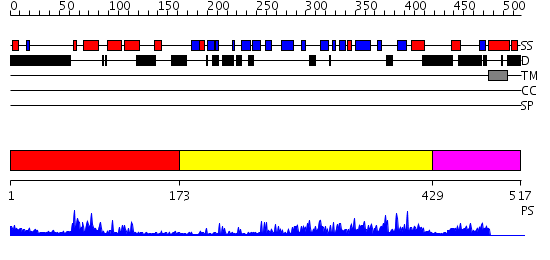
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..172] | 20.30103 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 2 | View Details | [173..428] | 46.0 | The crystal structure of Hsp40 Ydj1 | |
| 3 | View Details | [429..517] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|