













| General Information: |
|
| Name(s) found: |
MCM4 /
YPR019W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 933 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA pre-replicative complex [IDA cytoplasm [IDA MCM complex [IDA |
| Biological Process: |
DNA replication initiation
[TAS pre-replicative complex assembly [IPI DNA unwinding involved in replication [TAS |
| Molecular Function: |
chromatin binding
[TAS ATP-dependent DNA helicase activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQQSSSPTK EDNNSSSPVV PNPDSVPPQL SSPALFYSSS SSQGDIYGRN NSQNLSQGEG 60
61 NIRAAIGSSP LNFPSSSQRQ NSDVFQSQGR QGRIRSSASA SGRSRYHSDL RSDRALPTSS 120
121 SSLGRNGQNR VHMRRNDIHT SDLSSPRRIV DFDTRSGVNT LDTSSSSAPP SEASEPLRII 180
181 WGTNVSIQEC TTNFRNFLMS FKYKFRKILD EREEFINNTT DEELYYIKQL NEMRELGTSN 240
241 LNLDARNLLA YKQTEDLYHQ LLNYPQEVIS IMDQTIKDCM VSLIVDNNLD YDLDEIETKF 300
301 YKVRPYNVGS CKGMRELNPN DIDKLINLKG LVLRSTPVIP DMKVAFFKCN VCDHTMAVEI 360
361 DRGVIQEPAR CERIDCNEPN SMSLIHNRCS FADKQVIKLQ ETPDFVPDGQ TPHSISLCVY 420
421 DELVDSCRAG DRIEVTGTFR SIPIRANSRQ RVLKSLYKTY VDVVHVKKVS DKRLDVDTST 480
481 IEQELMQNKV DHNEVEEVRQ ITDQDLAKIR EVAAREDLYS LLARSIAPSI YELEDVKKGI 540
541 LLQLFGGTNK TFTKGGRYRG DINILLCGDP STSKSQILQY VHKITPRGVY TSGKGSSAVG 600
601 LTAYITRDVD TKQLVLESGA LVLSDGGVCC IDEFDKMSDS TRSVLHEVME QQTISIAKAG 660
661 IITTLNARSS ILASANPIGS RYNPNLPVTE NIDLPPPLLS RFDLVYLVLD KVDEKNDREL 720
721 AKHLTNLYLE DKPEHISQDD VLPVEFLTMY ISYAKEHIHP IITEAAKTEL VRAYVGMRKM 780
781 GDDSRSDEKR ITATTRQLES MIRLAEAHAK MKLKNVVELE DVQEAVRLIR SAIKDYATDP 840
841 KTGKIDMNLV QTGKSVIQRK LQEDLSREIM NVLKDQASDS MSFNELIKQI NEHSQDRVES 900
901 SDIQEALSRL QQEDKVIVLG EGVRRSVRLN NRV |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
CDC45 CDC6 DPB11 DPB2 DPB3 MCM2 MCM3 MCM4 MCM5 MCM6 MCM7 ORC1 ORC2 ORC3 ORC4 ORC5 ORC6 POL2 POL3 POL31 POL32 SLD3 |
| View Details | Ho Y, et al. (2002) |
|
ADR1 ARC15 ARC18 ARC19 ARC35 ARC40 ARP2 ARP3 BNI1 DUR1,2 IMH1 KAP122 MCM4 MCM7 MET18 RPA135 RPT4 SAN1 SRP1 TRP3 UBI4 YJR029W YPL113C |
| View Details | Qiu et al. (2008) |

|
ABF1 CDC45 CDC6 DPB11 DPB2 DPB3 DPB4 HCS1 MCM2 MCM3 MCM4 MCM5 MCM6 MCM7 MRC1 NOC3 ORC1 ORC2 ORC3 ORC4 ORC5 ORC6 POL1 POL2 POL3 POL30 POL31 POL32 RFA2 RFC1 RFC4 RFC5 RRM3 SLD3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
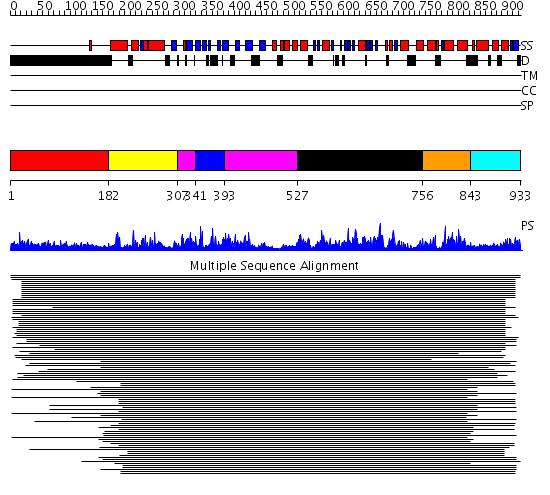
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..181] | 2.065996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [182..306] | 176.9691 | DNA replication initiator (cdc21/cdc54) N-terminal domain | |
| 3 | View Details | [307..340] [393..526] |
176.9691 | DNA replication initiator (cdc21/cdc54) N-terminal domain | |
| 4 | View Details | [341..392] | 176.9691 | DNA replication initiator (cdc21/cdc54) N-terminal domain | |
| 5 | View Details | [527..755] | 50.0 | ATPase subunit of magnesium chelatase, BchI | |
| 6 | View Details | [756..842] | 50.0 | ATPase subunit of magnesium chelatase, BchI | |
| 7 | View Details | [843..933] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)