













| General Information: |
|
| Name(s) found: |
DNL4 /
YOR005C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 944 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[TAS |
| Biological Process: |
double-strand break repair via nonhomologous end joining
[IDA |
| Molecular Function: |
DNA ligase (ATP) activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MISALDSIPE PQNFAPSPDF KWLCEELFVK IHEVQINGTA GTGKSRSFKY YEIISNFVEM 60
61 WRKTVGNNIY PALVLALPYR DRRIYNIKDY VLIRTICSYL KLPKNSATEQ RLKDWKQRVG 120
121 KGGNLSSLLV EEIAKRRAEP SSKAITIDNV NHYLDSLSGD RFASGRGFKS LVKSKPFLHC 180
181 VENMSFVELK YFFDIVLKNR VIGGQEHKLL NCWHPDAQDY LSVISDLKVV TSKLYDPKVR 240
241 LKDDDLSIKV GFAFAPQLAK KVNLSYEKIC RTLHDDFLVE EKMDGERIQV HYMNYGESIK 300
301 FFSRRGIDYT YLYGASLSSG TISQHLRFTD SVKECVLDGE MVTFDAKRRV ILPFGLVKGS 360
361 AKEALSFNSI NNVDFHPLYM VFDLLYLNGT SLTPLPLHQR KQYLNSILSP LKNIVEIVRS 420
421 SRCYGVESIK KSLEVAISLG SEGVVLKYYN SSYNVASRNN NWIKVKPEYL EEFGENLDLI 480
481 VIGRDSGKKD SFMLGLLVLD EEEYKKHQGD SSEIVDHSSQ EKHIQNSRRR VKKILSFCSI 540
541 ANGISQEEFK EIDRKTRGHW KRTSEVAPPA SILEFGSKIP AEWIDPSESI VLEIKSRSLD 600
601 NTETNMQKYA TNCTLYGGYC KRIRYDKEWT DCYTLNDLYE SRTVKSNPSY QAERSQLGLI 660
661 RKKRKRVLIS DSFHQNRKQL PISNIFAGLL FYVLSDYVTE DTGIRITRAE LEKTIVEHGG 720
721 KLIYNVILKR HSIGDVRLIS CKTTTECKAL IDRGYDILHP NWVLDCIAYK RLILIEPNYC 780
781 FNVSQKMRAV AEKRVDCLGD SFENDISETK LSSLYKSQLS LPPMGELEID SEVRRFPLFL 840
841 FSNRIAYVPR RKISTEDDII EMKIKLFGGK ITDQQSLCNL IIIPYTDPIL RKDCMNEVHE 900
901 KIKEQIKASD TIPKIARVVA PEWVDHSINE NCQVPEEDFP VVNY |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
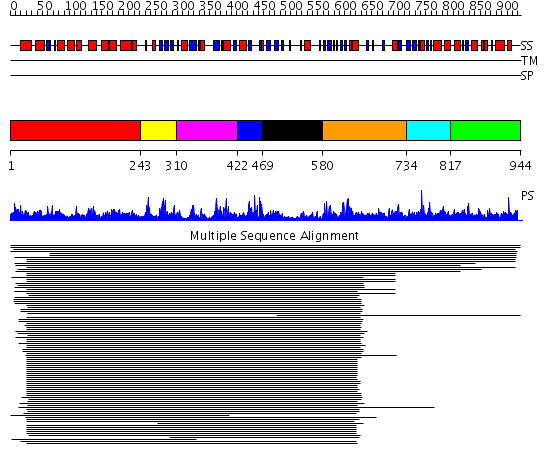
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..242] | 1.096977 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [243..309] | 8.04 | ATP-dependent DNA ligase; ATP-dependent DNA ligase, N-terminal domain | |
| 3 | View Details | [310..421] | 26.30103 | ATP-dependent DNA ligase; ATP-dependent DNA ligase, N-terminal domain | |
| 4 | View Details | [422..468] | 26.30103 | ATP-dependent DNA ligase; ATP-dependent DNA ligase, N-terminal domain | |
| 5 | View Details | [469..579] | 26.30103 | ATP-dependent DNA ligase; ATP-dependent DNA ligase, N-terminal domain | |
| 6 | View Details | [580..733] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [734..816] | 5.39794 | CRYSTAL STRUCTURE OF A XRCC4-DNA LIGASE IV COMPLEX | |
| 8 | View Details | [817..944] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)