













| General Information: |
|
| Name(s) found: |
UBP12 /
YJL197W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1254 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
ubiquitin-specific protease activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSSDVSSRE CSLVYNEDPD FTDGTTPCDR LGVDLMNVLD DKDEIKQESV PVSDREIEDT 60
61 ESDASAVSSF ASANELIAEP HAASETNLGT NGQDGRNVLE QQRDVVARLI EENKETQKEG 120
121 DKVCIVPKVW YDKFFDPDVT DPEDIGPINT RMICRDFENF VLEDYNRCPY LSIAEPVFNF 180
181 LSEIYGMTSG SYPVVTNLVI NQTTGELETE YNKWFFRLHY LTEKQDGRKR RHGQDDSIMY 240
241 LSMSALNLVR DLVEKSMNLF FEKADHLDVN AVDFKIWFVS EGSDIATDSN VSTFLNSSYE 300
301 ITPLQFLELP IKKLLIPDMF ENRLDKITSN PSDLVIEIKP IEGNHHWPSN YFAYNKLEPA 360
361 SGTTGLVNLG NTCYMNSALQ CLVHIPQLRD YFLYDGYEDE INEENPLGYH GYVARAFSDL 420
421 VQKLFQNRMS IMQRNAAFPP SMFKSTIGHF NSMFSGYMQQ DSQEFLAFLL DSLHEDLNRI 480
481 IKKEYTEKPS LSPGDDVNDW NVVKKLADDT WEMHLKRNCS VITDLFVGMY KSTLYCPECQ 540
541 NVSITFDPYN DVTLPLPVDT VWDKTIKIFP MNSPPLLLEV ELSKSSTYMD LKNYVGKMSG 600
601 LDPNTLFGCE IFSNQIYVNY ESTESNAQFL TLQELIKPAD DVIFYELPVT NDNEVIVPVL 660
661 NTRIEKGYKN AMLFGVPFFI TLKEDELNNP GAIRMKLQNR FVHLSGGYIP FPEPVGNRTD 720
721 FADAFPLLVE KYPDVEFEQY KDILQYTSIK VTDKDKSFFS IKILSVEKEQ QFASNNRTGP 780
781 NFWTPISQLN LDKATDIDDK LEDVVKDIYN YSSLVDCAEG VLMQVDDEGD TEGSEAKNFS 840
841 KPFQSGDDEE NKETVTNNEN VNNTNDRDED MELTDDVEED ASTEPELTDK PEALDKIKDS 900
901 LTSTPFAILS MNDIIVCEWS ELGSNEAFSD DKIYNWENPA TLPNKELENA KLERSNAKER 960
961 TITLDDCLQL FSKPEILGLT DSWYCPTCKE HRQATKQIQL WNTPDILLIH LKRFESQRSF1020
1021 SDKIDATVNF PITDLDLSRY VVYKDDPRGL IYDLYAVDNH YGGLGGGHYT AYVKNFADNK1080
1081 WYYFDDSRVT ETAPENSIAG SAYLLFYIRR HKDGNGLGSS KLQEIIQKSR HGYDERIKKI1140
1141 YDEQMKLYEF NKTDEEEDVS DDMIECNEDV QAPEYSNRSL EVGHIETQDC NDEDDNDDGE1200
1201 RTNSGRRKLR LLKKVYKNNS GLGSSSTSEI SEGCPENEVA DLNLKNGVTL ESPE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
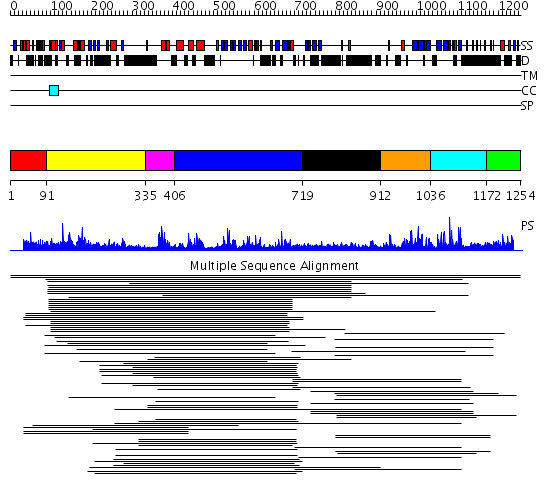
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..90] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [91..334] | 1.123954 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [335..405] | 18.958607 | No description for PF00442 was found. No confident structure predictions are available. | |
| 4 | View Details | [406..718] | 2.158982 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [719..911] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [912..1035] | 3.120991 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1036..1171] | 25.721246 | Ubiquitin carboxyl-terminal hydrolase No confident structure predictions are available. | |
| 8 | View Details | [1172..1254] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 |
|
|||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. | |||||||||
| 6 | No functions predicted. | |||||||||
| 7 | No functions predicted. | |||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)