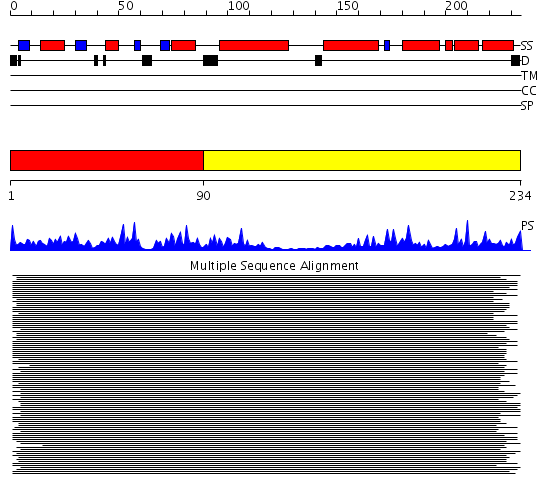
Description(s) found:
SHOW ONLY BEST
|
gi|731924|sp|P40582.1|GST1_YEAST RecName: Full=Glutathione S-transferase 1; AltName: Full=GST-I
[NCBI NR]
gi|6322229|ref|NP_012304.1| ER associated glutathione S-transferase capable of homodimerization; expression induced during the diauxic shift and throughout stationary phase; functional overlap with Gtt2p, Grx1p, and Grx2p; Gtt1p [Saccharomyces cerevisiae]
[NCBI NR]
pir||S48500 hypothetical protein YIR038c - yeast (Saccharomyces cerevisiae)
[NCBI NR]
gi|557844|emb|CAA86198.1| unnamed protein product [Saccharomyces cerevisiae]
[NCBI NR]
(P40582) Glutathione S-transferase I (EC 2.5.1.18) (GST-I)
[Swiss-Prot]
Glutathione S-transferase 1 OS=Saccharomyces cerevisiae GN=GTT1 PE=1 SV=1
[Swiss-Prot]
GTT1 Glutathione S-transferase
[mips]
ER associated glutathione S-transferase capable of homodimerization; expression induced during the diauxic shift and throughout stationary phase; functional overlap with Gtt2p, Grx1p, and Grx2p
[SGD]
GTT1, Chr IX from 423806-424510, reverse complement
[yeast_orfs2]
Chr IX from 423806-424510, reverse complement
[yeastorf3.fasta]
Chr IX from 424510-423806, reverse complement
[yeastorf5.fasta]
GTT1 SGDID:S0001477, Chr IX from 424510-423806, reverse complement, Verified ORF
[yeastflysequest.fasta]
ER associated glutathione S-transferase capable of homodimerization; expression induced during the diauxic shift and throughout stationary phase; functional overlap with Gtt2p, Grx1p, and Grx2p [Saccharomyces cerevisiae]�gi|557844|emb|CAA86198.1| unnamed
[nrdb_11152004_Chlamydomonas]
GTT1 SGDID:S000001477, Chr IX from 424510-423806, reverse complement, Verified ORF, "ER associated glutathione S-transferase capable of homodimerization; expression induced during the diauxic shift and throughout stationary phase; functional overlap with
[SGD_S-cerevisiae_na_12-16-2005_con_reversed.fasta]
unnamed protein product [Saccharomyces cerevisiae]
[nr-271106-contam.fasta]
Glutathione S-transferase I (GST-I)
[nr-271106-contam.fasta]
ER associated glutathione S-transferase capable of homodimerization; expression induced during the diauxic shift and throughout stationary phase; functional overlap with Gtt2p, Grx1p, and Grx2p; Gtt1p [Saccharomyces cerevisiae]
[nr-271106-contam.fasta]
GTT1 SGDID:S000001477, Chr IX from 424510-423806, reverse complement, Verified ORF, "ER associated glutathione S-transferase capable of homodimerization; expression induced during the diauxic shift and throughout stationary phase; functional overlap with Gtt2p, Grx1p, and Grx2p"
[yeast-bovrabprots.fasta]
[chemostatdb.fasta]
Glutathione S-transferase 1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) GN=GTT1 PE=1 SV=1
[UniProt_S_cerevisiae_20120516_05-16-2012_reversed.fasta]
GTT1 SGDID:S000001477, Chr IX from 424513-423809, Genome Release 64-1-1, reverse complement, Verified ORF, "ER associated glutathione S-transferase capable of homodimerization; expression induced during the diauxic shift and throughout stationary phase; functional overlap with Gtt2p, Grx1p, and Grx2p"
[yeast-030211-contam.fasta]
GTT1 SGDID:S000001477, Chr IX from 424513-423809, Genome Release 64-2-1, reverse complement, Verified ORF, "ER associated glutathione S-transferase capable of homodimerization; expression induced during the diauxic shift and throughout stationary phase; functional overlap with Gtt2p, Grx1p, and Grx2p"
[20141208_orf_trans.fasta]
YIR038C SGDID:S000001477, Chr IX from 424513-423809, Genome Release 64-1-1, reverse complement, Verified ORF, "ER associated glutathione S-transferase capable of homodimerization; expression induced during the diauxic shift and throughout stationary phase; functional overlap with Gtt2p, Grx1p, and Grx2p"
[2016-01-SGD_Yeast_Contam_Riffle_20140507-Ska-Hec-110-tubulin.fasta]
|