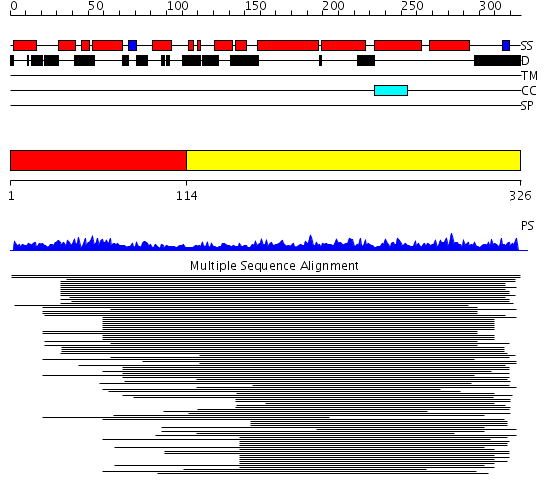
Description(s) found:
SHOW ONLY BEST
|
gi|731801|sp|P40531.1|GVP36_YEAST RecName: Full=Protein GVP36; AltName: Full=36 kDa Golgi vesicle protein
[NCBI NR]
gi|6322148|ref|NP_012223.1| Golgi vesicle protein of unknown function; localizes to both early and late Golgi vesicles; may interact with ribosomes, based on co-purification experiments; Gvp36p [Saccharomyces cerevisiae]
[NCBI NR]
gi|600007|emb|CAA86910.1| unknown [Saccharomyces cerevisiae]
[NCBI NR]
pir||S49937 hypothetical protein YIL041w - yeast (Saccharomyces cerevisiae)
[NCBI NR]
(P40531) 36.7 kDa protein in CBR5-NOT3 intergenic region
[Swiss-Prot]
Protein GVP36 OS=Saccharomyces cerevisiae GN=GVP36 PE=1 SV=1
[Swiss-Prot]
GVP36 Golgi-vesicle protein of unknown function
[mips]
BAR domain-containing protein that localizes to both early and late Golgi vesicles; required for adaptation to varying nutrient concentrations, fluid-phase endocytosis, polarization of the actin cytoskeleton, and vacuole biogenesis
[SGD]
YIL041W, Chr IX from 276524-277504
[yeast_orfs2]
Chr IX from 276524-277504
[yeastorf3.fasta]
GVP36 SGDID:S0001303, Chr IX from 276524-277504, Uncharacterized ORF
[yeastflysequest.fasta]
Golgi-vesicle protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm [Saccharomyces cerevisiae]�gi|600007|emb|CAA86910.1| unknown [Saccharomyces cerevisiae]�gi|731801|sp|P40531|YIE1_YEAST 36.7 kDa protein in
[nrdb_11152004_Chlamydomonas]
GVP36 SGDID:S000001303, Chr IX from 276524-277504, Uncharacterized ORF, "Golgi vesicle protein of unknown function; localizes to both early and late Golgi vesicles"
[SGD_S-cerevisiae_na_12-16-2005_con_reversed.fasta]
GVP36 SGDID:S000001303, Chr IX from 276524-277504, Verified ORF, "BAR domain-containing protein that localizes to both early and late Golgi vesicles; required for adaptation to varying nutrient concentrations, fluid-phase endocytosis, polarization of the
[SGD_S-cerevisiae_na_08-19-2008_con_reversed.fasta]
unknown [Saccharomyces cerevisiae]
[nr-271106-contam.fasta]
Golgi vesicle protein of unknown function; localizes to both early and late Golgi vesicles; Gvp36p [Saccharomyces cerevisiae]
[nr-271106-contam.fasta]
Protein GVP36 (36 kDa Golgi vesicle protein)
[nr-271106-contam.fasta]
GVP36 SGDID:S000001303, Chr IX from 276524-277504, Verified ORF, "BAR domain-containing protein that localizes to both early and late Golgi vesicles; required for adaptation to varying nutrient concentrations, fluid-phase endocytosis, polarization of the actin cytoskeleton, and vacuole biogenesis"
[yeast-bovrabprots.fasta]
[chemostatdb.fasta]
Protein GVP36 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) GN=GVP36 PE=1 SV=1
[UniProt_S_cerevisiae_20120516_05-16-2012_reversed.fasta]
GVP36 SGDID:S000001303, Chr IX from 276525-277505, Genome Release 64-1-1, Verified ORF, "BAR domain-containing protein that localizes to both early and late Golgi vesicles; required for adaptation to varying nutrient concentrations, fluid-phase endocytosis, polarization of the actin cytoskeleton, and vacuole biogenesis"
[yeast-030211-contam.fasta]
GVP36 SGDID:S000001303, Chr IX from 276525-277505, Genome Release 64-2-1, Verified ORF, "BAR domain protein that localizes to early and late Golgi vesicles; required for adaptation to varying nutrient concentrations, fluid-phase endocytosis, polarization of the actin cytoskeleton, and vacuole biogenesis"
[20141208_orf_trans.fasta]
TR5386|c0_g1_i1|g.3771 ORF TR5386|c0_g1_i1|g.3771 TR5386|c0_g1_i1|m.3771 type:complete len:327 (+) TR5386|c0_g1_i1:29-1009(+)
[Y12_S_cerevisiae-Sep2015-contam.fasta]
TR4977|c0_g1_i1|g.3317 ORF TR4977|c0_g1_i1|g.3317 TR4977|c0_g1_i1|m.3317 type:complete len:327 (+) TR4977|c0_g1_i1:29-1009(+)
[DBVPG6044_S_cerevisiae-Sep2015-contam.fasta]
YIL041W SGDID:S000001303, Chr IX from 276525-277505, Genome Release 64-1-1, Verified ORF, "BAR domain-containing protein that localizes to both early and late Golgi vesicles; required for adaptation to varying nutrient concentrations, fluid-phase endocytosis, polarization of the actin cytoskeleton, and vacuole biogenesis"
[2016-01-SGD_Yeast_Contam_Riffle_20140507-Ska-Hec-110-tubulin.fasta]
asmbl_8789|g.6392 ORF asmbl_8789|g.6392 asmbl_8789|m.6392 type:complete len:327 (+) asmbl_8789:42-1022(+)
[DBVPG6044_Trinity_v2_2_0_PASA_6May2016-contam.fasta]
asmbl_8926|g.6509 ORF asmbl_8926|g.6509 asmbl_8926|m.6509 type:complete len:327 (+) asmbl_8926:36-1016(+)
[Y12_Trinity_v2_2_0_PASA_6May2016-contam.fasta]
ID=asmbl_8762|g.5486 ORF ID=asmbl_8762|g.5486 ID=asmbl_8762|m.5486 type:complete len:327 (+) ID=asmbl_8762:42-1022(+)
[DBVPG6044_quiverGenomes_Trinity_v2_2_0_PASA_2June2016-contam.fasta]
ID=asmbl_8932|g.5370 ORF ID=asmbl_8932|g.5370 ID=asmbl_8932|m.5370 type:complete len:327 (+) ID=asmbl_8932:36-1016(+)
[Y12_quiver_Trinity_v2_2_0_PASA_2June2016-contam.fasta]
|