













| General Information: |
|
| Name(s) found: |
MDJ1 /
YFL016C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 511 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA mitochondrial inner membrane [IDA |
| Biological Process: |
proteolysis
[IMP protein folding [IDA |
| Molecular Function: |
chaperone binding
[IMP unfolded protein binding [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAFQQGVLSR CSGVFRHHVG HSRHINNILY RHAIAFASIA PRIPKSSFHT SAIRNNEAFK 60
61 DPYDTLGLKK SATGAEIKKA YYKLAKKYHP DINKEPDAEK KFHDLQNAYE ILSDETKRQQ 120
121 YDQFGPAAFG GGGAAGGAGG GSGSPFGSQF HDFSGFTSAG GSPFGGINFE DLFGAAFGGG 180
181 GRGSGGASRS SSMFRQYRGD PIEIVHKVSF KDAVFGSKNV QLRFSALDPC STCSGTGMKP 240
241 NTHKVSCSTC HGTGTTVHIR GGFQMMSTCP TCNGEGTMKR PQDNCTKCHG EGVQVNRAKT 300
301 ITVDLPHGLQ DGDVVRIPGQ GSYPDIAVEA DLKDSVKLSR GDILVRIRVD KDPNFSIKNK 360
361 YDIWYDKEIP ITTAALGGTV TIPTVEGQKI RIKVAPGTQY NQVISIPNMG VPKTSTIRGD 420
421 MKVQYKIVVK KPQSLAEKCL WEALADVTND DMAKKTMQPG TAAGTAINEE ILKKQKQEEE 480
481 KHAKKDDDNT LKRLENFITN TFRKIKGDKK N |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
MDJ1 RPL11B |
| View Details | Krogan NJ, et al. (2006) |
|
MDJ1 SCC2 SCC4 |
| View Details | Ho Y, et al. (2002) |
|
ACC1 CCT6 CDC33 CPR6 DMC1 DPM1 FMP10 MDJ1 MTC5 NOP14 NPR2 POX1 RNQ1 RVB2 SEH1 THI21 TRP3 YHR033W |
| View Details | Qiu et al. (2008) |

|
HSP78 MDJ1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
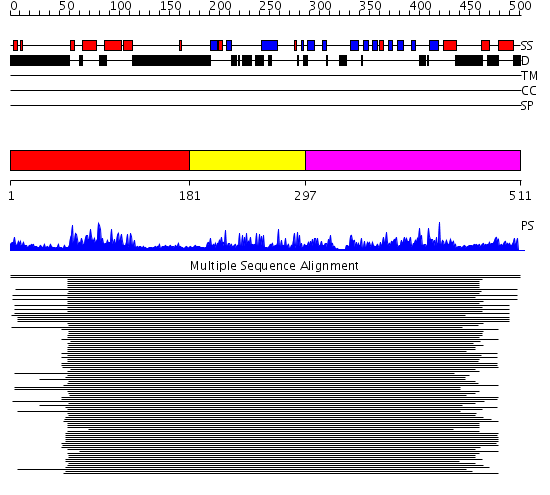
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..180] | 169.0 | DnaJ chaperone, N-terminal (J) domain | |
| 2 | View Details | [181..296] | 142.64593 | Cysteine-rich domain of the chaperone protein DnaJ. | |
| 3 | View Details | [297..511] | 22.30103 | Heat shock protein 40 Sis1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.68 |
Source: Reynolds et al. (2008)