













| General Information: |
|
| Name(s) found: |
MET6 /
YER091C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 767 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
methionine biosynthetic process
[IMP |
| Molecular Function: |
5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVQSAVLGFP RIGPNRELKK ATEGYWNGKI TVDELFKVGK DLRTQNWKLQ KEAGVDIIPS 60
61 NDFSFYDQVL DLSLLFNVIP DRYTKYDLSP IDTLFAMGRG LQRKATETEK AVDVTALEMV 120
121 KWFDSNYHYV RPTFSKTTQF KLNGQKPVDE FLEAKELGIH TRPVLLGPVS YLFLGKADKD 180
181 SLDLEPLSLL EQLLPLYTEI LSKLASAGAT EVQIDEPVLV LDLPANAQAA IKKAYTYFGE 240
241 QSNLPKITLA TYFGTVVPNL DAIKGLPVAA LHVDFVRAPE QFDEVVAAIG NKQTLSVGIV 300
301 DGRNIWKNDF KKSSAIVNKA IEKLGADRVV VATSSSLLHT PVDLNNETKL DAEIKGFFSF 360
361 ATQKLDEVVV ITKNVSGQDV AAALEANAKS VESRGKSKFI HDAAVKARVA SIDEKMSTRA 420
421 APFEQRLPEQ QKVFNLPLFP TTTIGSFPQT KDIRINRNKF NKGTISAEEY EKFINSEIEK 480
481 VIRFQEEIGL DVLVHGEPER NDMVQYFGEQ INGYAFTVNG WVQSYGSRYV RPPIIVGDLS 540
541 RPKAMSVKES VYAQSITSKP VKGMLTGPIT CLRWSFPRDD VDQKTQAMQL ALALRDEVND 600
601 LEAAGIKVIQ VDEPALREGL PLREGTERSA YYTWAAEAFR VATSGVANKT QIHSHFCYSD 660
661 LDPNHIKALD ADVVSIEFSK KDDANYIAEF KNYPNHIGLG LFDIHSPRIP SKDEFIAKIS 720
721 TILKSYPAEK FWVNPDCGLK TRGWEETRLS LTHMVEAAKY FREQYKN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
ALA1 ARO1 DUG1 GDH1 HIS4 HIS6 HRD1 MET6 OYE2 RAD14 SCP160 SPE3 SPE4 TIM11 URA6 YLF2 YRB2 |
| View Details | Gavin AC, et al. (2006) |

|
MET6 TKL1 |
| View Details | Ho Y, et al. (2002) |
|
AAT2 ACH1 ACO1 AFR1 AHP1 ARC1 ARF1 ATP3 CAR2 CCT6 CDC12 CDC14 CDC33 CLU1 COF1 COR1 CPR1 CYR1 CYS4 DOG1 DOG2 EGD1 EGD2 ERB1 ERG13 ERG6 FPR1 FRS2 GND1 GND2 GRX1 GUS1 HAS1 HEF3 HHF1, HHF2 HXT7 IMD4 LRO1 LSP1 MET6 MRT4 MSK1 MUB1 NOC2 NOC3 NOG1 NSA1 NTF2 NUG1 OLA1 OYE2 PFK1 PIL1 PRM2 PRP43 PYC1 RGD1 RNR2 RSN1 SAH1 SCP160 SCS2 SEC53 SEN15 SES1 SNU13 SOD1 SPB4 SSQ1 TEF4 THS1 TIF1, TIF2 TIF6 TMA19 TSA2 UBA1 UBR2 VMA22 VMA4 VMA5 WTM1 YNK1 YTM1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
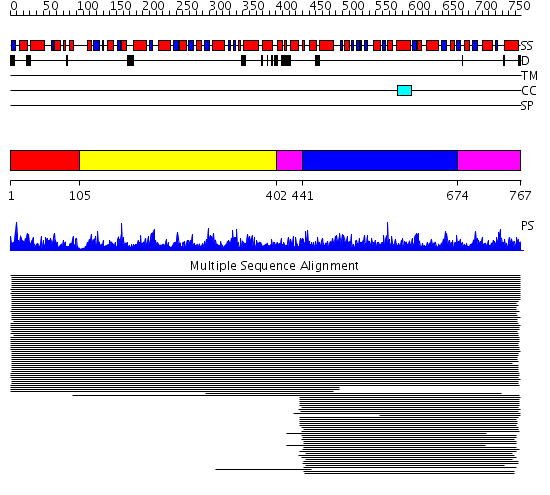
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..104] | 1.042975 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [105..401] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [402..440] [674..767] |
11.96 | Uroporphyrinogen decarboxylase, UROD | |
| 4 | View Details | [441..673] | 11.96 | Uroporphyrinogen decarboxylase, UROD |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)