













| General Information: |
|
| Name(s) found: |
LCD1 /
YDR499W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 747 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear chromosome
[IDA |
| Biological Process: |
establishment of protein localization
[IDA DNA damage checkpoint [IMP telomere maintenance via telomerase [IMP |
| Molecular Function: |
protein binding
[IDA damaged DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRRETVGEFS SDDDDDILLE LGTRPPRFTQ IPPSSAALQT QIPTTLEVTT TTLNNKQSKN 60
61 DNQLVNQLNK AQGEASMLRD KINFLNIERE KEKNIQAVKV NELQVKHLQE LAKLKQELQK 120
121 LEDEKKFLQM EARGKSKREV ITNVKPPSTT LSTNTNTITP DSSSVAIEAK PQSPQSKKRK 180
181 ISDNLLKKNM VPLNPNRIIP DETSLFLESI LLHQIIGADL STIEILNRLK LDYITEFKFK 240
241 NFVIAKGAPI GKSIVSLLLR CKKTLTLDRF IDTLLEDIAV LIKEISVHPN ESKLAVPFLV 300
301 ALMYQIVQFR PSATHNLALK DCFLFICDLI RIYHHVLKVP IHESNMNLHV EPQIFQYELI 360
361 DYLIISYSFD LLEGILRVLQ SHPKQTYMEF FDENILKSFE FVYKLALTIS YKPMVNVIFS 420
421 AVEVVNIITS IILNMDNSSD LKSLISGSWW RDCITRLYAL LEKEIKSGDV YNENVDTTTL 480
481 HMSKYHDFFG LIRNIGDNEL GGLISKLIYT DRLQSVPRVI SKEDIGMDSD KFTAPIIGYK 540
541 MEKWLLKLKD EVLNIFENLL MIYGDDATIV NGEMLIHSSK FLSREQALMI ERYVGQDSPN 600
601 LDLRCHLIEH TLTIIYRLWK DHFKQLREEQ IKQVESQLIM SLWRFLVCQT ETVTANEREM 660
661 RDHRHLVDSL HDLTIKDQAS YYEDAFEDLP EYIEEELKMQ LNKRTGRIMQ VKYDEKFQEM 720
721 ARTILESKSF DLTTLEEADS LYISMGL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
LCD1 VAC17 |
| View Details | Gavin AC, et al. (2002) |

|
CMR1 DNA2 HDA1 HDA2 HDA3 HSC82 KAP95 LCD1 MEC1 MGM101 MLH1 MPH1 MSH2 MSH3 MSH6 OLA1 PGK1 PMS1 PRO2 RAD16 RAD52 RFA1 RFA2 RFA3 RIM1 RNH1 RSC8 RVB2 SGS1 SHS1 TIF1, TIF2 TOP3 VAS1 VPS1 YHR122W |
| View Details | Ho Y, et al. (2002) |
|
ADH2 ADH5 HEM15 HHF1, HHF2 ILV5 LCD1 RNQ1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
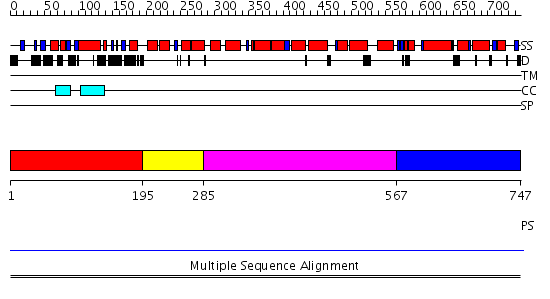
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..194] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [195..284] | 8.0 | CBP80, 80KDa nuclear cap-binding protein | |
| 3 | View Details | [285..566] | 8.0 | CBP80, 80KDa nuclear cap-binding protein | |
| 4 | View Details | [567..747] | 7.09 | Hemolysin E (HlyE, ClyA, SheA) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)