













| General Information: |
|
| Name(s) found: |
PPN1 /
YDR452W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 674 amino acids |
Gene Ontology: |
|
| Cellular Component: |
vacuolar membrane
[IDA |
| Biological Process: |
polyphosphate metabolic process
[IDA |
| Molecular Function: |
endopolyphosphatase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVVVGKSEVR NVSMSRPKKK SLIAILSTCV LFFLVFIIGA KFQYVSVFSK FLDDRGDNES 60
61 LQLLNDIEFT RLGLTPREPV IIKDVKTGKE RKLHGRFLHI TDIHPDPYYV EGSSIDAVCH 120
121 TGKPSKKKDV APKFGKAMSG CDSPVILMEE TLRWIKENLR DKIDFVIWTG DNIRHDNDRK 180
181 HPRTEAQIFD MNNIVADKMT ELFSAGNEED PRDFDVSVIP SLGNNDVFPH NMFALGPTLQ 240
241 TREYYRIWKN FVPQQQQRTF DRSASFLTEV IPGKLAVLSI NTLYLFKANP LVDNCNSKKE 300
301 PGYQLLLWFG YVLEELRSRG MKVWLSGHVP PIAKNFDQSC YDKFTLWTHE YRDIIIGGLY 360
361 GHMNIDHFIP TDGKKARKSL LKAMEQSTRV QQGEDSNEED EETELNRILD HAMAAKEVFL 420
421 MGAKPSNKEA YMNTVRDTYY RKVWNKLERV DEKNVENEKK KKEKKDKKKK KPITRKELIE 480
481 RYSIVNIGGS VIPTFNPSFR IWEYNITDIV NDSNFAVSEY KPWDEFFESL NKIMEDSLLE 540
541 DEMDSSNIEV GINREKMGEK KNKKKKKNDK TMPIEMPDKY ELGPAYVPQL FTPTRFVQFY 600
601 ADLEKINQEL HNSFVESKDI FRYEIEYTSD EKPYSMDSLT VGSYLDLAGR LYENKPAWEK 660
661 YVEWSFASSG YKDD |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | sample: hx4 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | hx23: mixture in detergent (control) Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
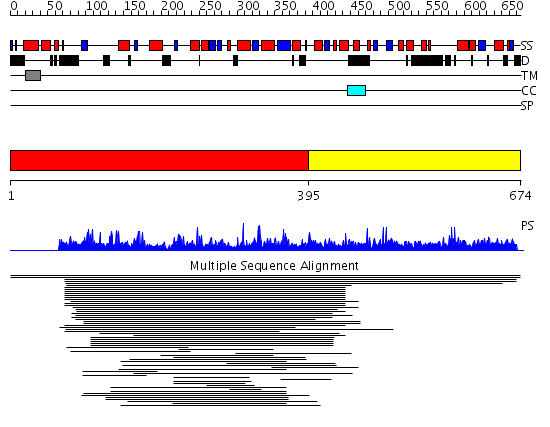
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..394] | 13.8 | Mammalian purple acid phosphatase | |
| 2 | View Details | [395..674] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|