













| General Information: |
|
| Name(s) found: |
HO /
YDL227C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 586 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[TAS |
| Biological Process: |
gene conversion at mating-type locus, DNA double-strand break formation
[IDA mating type switching [IMP gene conversion at mating-type locus [TAS |
| Molecular Function: |
endonuclease activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSENTTILM ANGEIKDIAN VTANSYVMCA DGSAARVINV TQGYQKIYNI QQKTKHRAFE 60
61 GEPGRLDPRR RTVYQRLALQ CTAGHKLSVR VPTKPLLEKS GRNATKYKVR WRNLQQCQTL 120
121 DGRIIIIPKN HHKTFPMTVE GEFAAKRFIE EMERSKGEYF NFDIEVRDLD YLDAQLRISS 180
181 CIRFGPVLAG NGVLSKFLTG RSDLVTPAVK SMAWMLGLWL GDSTTKEPEI SVDSLDPKLM 240
241 ESLRENAKIW GLYLTVCDDH VPLRAKHVRL HYGDGPDENR KTRNLRKNNP FWKAVTILKF 300
301 KRDLDGEKQI PEFMYGEHIE VREAFLAGLI DSDGYVVKKG EGPESYKIAI QTVYSSIMDG 360
361 IVHISRSLGM SATVTTRSAR EEIIEGRKVQ CQFTYDCNVA GGTTSQNVLS YCRSGHKTRE 420
421 VPPIIKREPV YFSFTDDFQG ESTVYGLTIE GHKNFLLGNK IEVKSCRGCC VGEQLKISQK 480
481 KNLKHCVACP RKGIKYFYKD WSGKNRVCAR CYGRYKFSGH HCINCKYVPE AREVKKAKDK 540
541 GEKLGITPEG LPVKGPECIK CGGILQFDAV RGPHKSCGNN AGARIC |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
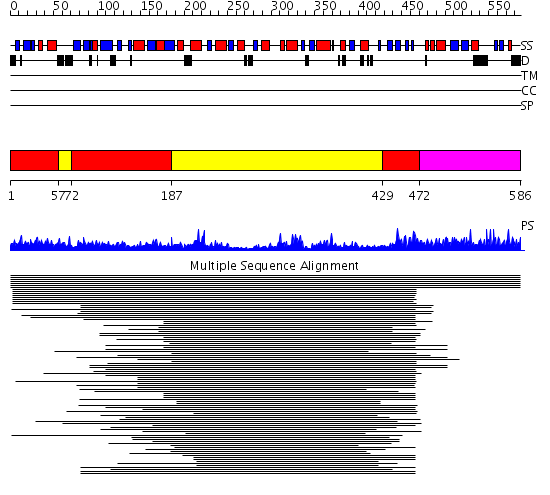
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..56] [72..186] [429..471] |
634.221849 | VMA1-derived endonuclease (VDE) PI-Scei intein; VMA1-derived endonuclease (VDE) PI-SceI | |
| 2 | View Details | [57..71] [187..428] |
634.221849 | VMA1-derived endonuclease (VDE) PI-Scei intein; VMA1-derived endonuclease (VDE) PI-SceI | |
| 3 | View Details | [472..586] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)