













| General Information: |
|
| Name(s) found: |
PGK1 /
YCR012W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 416 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA |
| Biological Process: |
gluconeogenesis
[IMP glycolysis [IMP |
| Molecular Function: |
phosphoglycerate kinase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLSSKLSVQ DLDLKDKRVF IRVDFNVPLD GKKITSNQRI VAALPTIKYV LEHHPRYVVL 60
61 ASHLGRPNGE RNEKYSLAPV AKELQSLLGK DVTFLNDCVG PEVEAAVKAS APGSVILLEN 120
121 LRYHIEEEGS RKVDGQKVKA SKEDVQKFRH ELSSLADVYI NDAFGTAHRA HSSMVGFDLP 180
181 QRAAGFLLEK ELKYFGKALE NPTRPFLAIL GGAKVADKIQ LIDNLLDKVD SIIIGGGMAF 240
241 TFKKVLENTE IGDSIFDKAG AEIVPKLMEK AKAKGVEVVL PVDFIIADAF SADANTKTVT 300
301 DKEGIPAGWQ GLDNGPESRK LFAATVAKAK TIVWNGPPGV FEFEKFAAGT KALLDEVVKS 360
361 SAAGNTVIIG GGDTATVAKK YGVTDKISHV STGGGASLEL LEGKELPGVA FLSEKK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ENO2 FBA1 PGK1 |
| View Details | Gavin AC, et al. (2006) |
|
CHS1 ENO2 PGK1 |
| View Details | Gavin AC, et al. (2002) |
|
ADA2 ADH1 BLM10 CDC6 CMR1 COG3 COP1 CSE2 DNA2 ECM29 ENO2 FAB1 FBA1 GAL11 GCN5 GFA1 GLO3 GPT2 HDA1 HDA2 HDA3 HSC82 ILV1 KAP95 LCD1 MEC1 MED1 MED11 MED2 MED4 MED6 MED7 MED8 MGM101 MLH1 MLH2 MLP1 MPH1 MRPL6 MRPS28 MSH2 MSH3 MSH6 NAS6 NUT1 NUT2 OLA1 PGD1 PGK1 PMS1 PRE1 PRE10 PRE2 PRE3 PRE4 PRE5 PRE6 PRE7 PRE8 PRE9 PRO2 PUP3 RAD16 RAD23 RAD34 RAD4 RAD52 RET2 RET3 RFA1 RFA2 RFA3 RGR1 RIM1 RNH1 ROX3 RPG1 RPN10 RPN11 RPN12 RPN13 RPN3 RPN5 RPN6 RPN7 RPN8 RPN9 RPO21 RPT1 RPT2 RPT3 RPT4 RPT5 RPT6 RSC8 RVB2 SAM1 SCJ1 SCL1 SEC21 SEC26 SEC27 SEC28 SEH1 SGS1 SHS1 SIN4 SPF1 SPT7 SRB2 SRB4 SRB5 SRB6 SRB7 SRB8 SRO7 SRP1 SSE2 SSL1 SSN2 SSN3 SSN8 TAF6 TFA1 TFB1 TFB4 TIF1, TIF2 TOP3 TOS1 UBP6 VAS1 VPS1 YFL006W YHR122W YIL077C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | McCusker D, et al (2007) | |
| View Run | No Comments | Widlund PO, et al. (2005) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | experimental2 - mih1-2 | McCusker D, et al (2007) | |
| View Run | 3912: TAP-tagged, strain background includes hpc2(delta) | Green EM, et al (2005) | |
| View Run | cac1: TAP-tagged | Green EM, et al (2005) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #22 Asynchronous Prep2-IMAC Phosphopeptide enrichment B1-2 | Keck JM, et al. (2011) | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
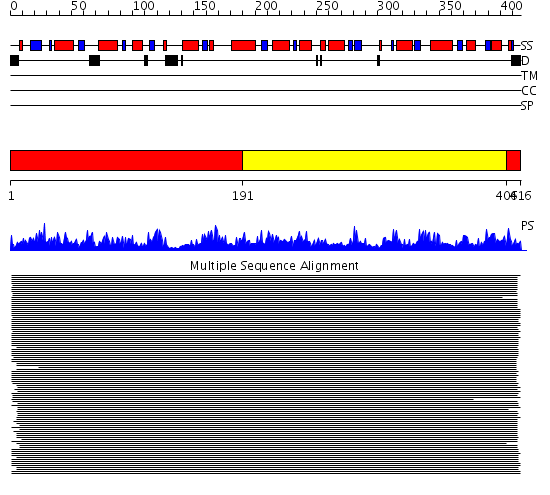
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..190] [406..416] |
11000.0 | Phosphoglycerate kinase | |
| 2 | View Details | [191..405] | 11000.0 | Phosphoglycerate kinase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)