













| General Information: |
|
| Name(s) found: |
CDC27 /
YBL084C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 758 amino acids |
Gene Ontology: |
|
| Cellular Component: |
anaphase-promoting complex
[TAS |
| Biological Process: |
mitotic spindle elongation
[TAS cyclin catabolic process [TAS mitotic sister chromatid segregation [TAS anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process [IDA mitotic metaphase/anaphase transition [TAS protein ubiquitination [IPI |
| Molecular Function: |
ubiquitin-protein ligase activity
[TAS protein binding [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAVNPELAPF TLSRGIPSFD DQALSTIIQL QDCIQQAIQQ LNYSTAEFLA ELLYAECSIL 60
61 DKSSVYWSDA VYLYALSLFL NKSYHTAFQI SKEFKEYHLG IAYIFGRCAL QLSQGVNEAI 120
121 LTLLSIINVF SSNSSNTRIN MVLNSNLVHI PDLATLNCLL GNLYMKLDHS KEGAFYHSEA 180
181 LAINPYLWES YEAICKMRAT VDLKRVFFDI AGKKSNSHNN NAASSFPSTS LSHFEPRSQP 240
241 SLYSKTNKNG NNNINNNVNT LFQSSNSPPS TSASSFSSIQ HFSRSQQQQA NTSIRTCQNK 300
301 NTQTPKNPAI NSKTSSALPN NISMNLVSPS SKQPTISSLA KVYNRNKLLT TPPSKLLNND 360
361 RNHQNNNNNN NNNNNNNNNN NNNNNNNNII NKTTFKTPRN LYSSTGRLTT SKKNPRSLII 420
421 SNSILTSDYQ ITLPEIMYNF ALILRSSSQY NSFKAIRLFE SQIPSHIKDT MPWCLVQLGK 480
481 LHFEIINYDM SLKYFNRLKD LQPARVKDME IFSTLLWHLH DKVKSSNLAN GLMDTMPNKP 540
541 ETWCCIGNLL SLQKDHDAAI KAFEKATQLD PNFAYAYTLQ GHEHSSNDSS DSAKTCYRKA 600
601 LACDPQHYNA YYGLGTSAMK LGQYEEALLY FEKARSINPV NVVLICCCGG SLEKLGYKEK 660
661 ALQYYELACH LQPTSSLSKY KMGQLLYSMT RYNVALQTFE ELVKLVPDDA TAHYLLGQTY 720
721 RIVGRKKDAI KELTVAMNLD PKGNQVIIDE LQKCHMQE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
APC1 APC2 APC4 CDC16 CDC23 CDC27 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
APC1 APC11 APC2 APC4 APC5 APC9 CDC16 CDC23 CDC26 CDC27 DOC1 |
| View Details | Krogan NJ, et al. (2006) |
|
APC1 APC2 APC4 APC5 CDC16 CDC23 CDC26 CDC27 DOC1 FMP52 MND2 SSP2 |
| View Details | Gavin AC, et al. (2006) |
|
APC1 APC2 APC4 APC5 APC9 CDC16 CDC23 CDC27 |
| View Details | Gavin AC, et al. (2002) |
|
ACT1 APC1 APC2 CDC16 CDC23 CDC27 |
| View Details | Qiu et al. (2008) |
|
APC1 APC11 APC2 APC4 APC5 APC9 CDC16 CDC23 CDC26 CDC27 DOC1 MND2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
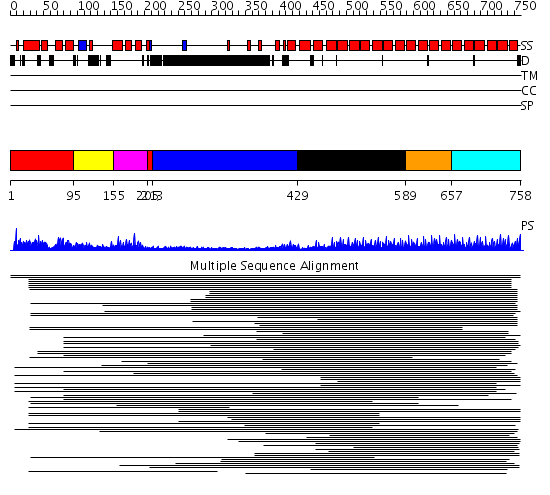
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..94] [205..212] |
8.93 | Neutrophil cytosolic factor 2 (NCF-2, p67-phox) | |
| 2 | View Details | [95..154] | 8.93 | Neutrophil cytosolic factor 2 (NCF-2, p67-phox) | |
| 3 | View Details | [155..204] | 8.93 | Neutrophil cytosolic factor 2 (NCF-2, p67-phox) | |
| 4 | View Details | [213..428] | 7.73 | No description for 1ldvA was found. | |
| 5 | View Details | [429..588] | 122.9794 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 6 | View Details | [589..656] | 122.9794 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 7 | View Details | [657..758] | 122.9794 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)