













| General Information: |
|
| Name(s) found: |
RECE_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 866 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTKPLFLLR KAKKSSGEPD VVLWASNDFE STCATLDYLI VKSGKKLSSY FKAVATNFPV 60
61 VNDLPAEGEI DFTWSERYQL SKDSMTWELK PGAAPDNAHY QGNTNVNGED MTEIEENMLL 120
121 PISGQELPIR WLAQHGSEKP VTHVSRDGLQ ALHIARAEEL PAVTALAVSH KTSLLDPLEI 180
181 RELHKLVRDT DKVFPNPGNS NLGLITAFFE AYLNADYTDR GLLTKEWMKG NRVSHITRTA 240
241 SGANAGGGNL TDRGEGFVHD LTSLARDVAT GVLARSMDLD IYNLHPAHAK RIEEIIAENK 300
301 PPFSVFRDKF ITMPGGLDYS RAIVVASVKE APIGIEVIPA HVTEYLNKVL TETDHANPDP 360
361 EIVDIACGRS SAPMPQRVTE EGKQDDEEKP QPSGTTAVEQ GEAETMEPDA TEHHQDTQPL 420
421 DAQSQVNSVD AKYQELRAEL HEARKNIPSK NPVDDDKLLA ASRGEFVDGI SDPNDPKWVK 480
481 GIQTRDCVYQ NQPETEKTSP DMNQPEPVVQ QEPEIACNAC GQTGGDNCPD CGAVMGDATY 540
541 QETFDEESQV EAKENDPEEM EGAEHPHNEN AGSDPHRDCS DETGEVADPV IVEDIEPGIY 600
601 YGISNENYHA GPGISKSQLD DIADTPALYL WRKNAPVDTT KTKTLDLGTA FHCRVLEPEE 660
661 FSNRFIVAPE FNRRTNAGKE EEKAFLMECA STGKTVITAE EGRKIELMYQ SVMALPLGQW 720
721 LVESAGHAES SIYWEDPETG ILCRCRPDKI IPEFHWIMDV KTTADIQRFK TAYYDYRYHV 780
781 QDAFYSDGYE AQFGVQPTFV FLVASTTIEC GRYPVEIFMM GEEAKLAGQQ EYHRNLRTLS 840
841 DCLNTDEWPA IKTLSLPRWA KEYAND |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
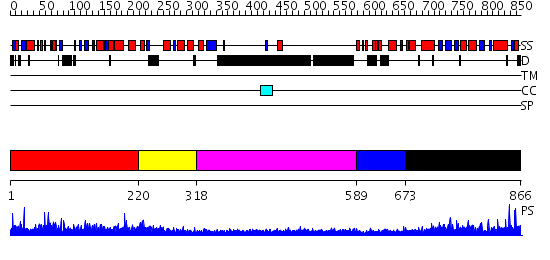
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..219] | 134.318759 | No description for PF06630.2 was found. No confident structure predictions are available. | |
| 2 | View Details | [220..317] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [318..588] | 1.280996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [589..672] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [673..866] | 3.198988 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)