













| General Information: |
|
| Name(s) found: |
gi|7269087
[NCBI NR]
gi|5738372 [NCBI NR] gi|25407585 [NCBI NR] gi|15235641 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 340 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endomembrane system
[IEA]
intracellular [IEA] |
| Biological Process: |
ubiquitin-dependent protein catabolic process
[IEA][ISS]
|
| Molecular Function: |
zinc ion binding
[IEA]
ubiquitin thiolesterase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVEFQVLDFH FERFCSVSLS NLNVYACLVC GKYFQGRSQK SHAYTHSLEA GHHVYINLLT 60
61 EKVYCLPDSY EINDPSLDDI RHVLNPRFSR AQVNELDKNR QWSRALDGSD YLPGMVGLNN 120
121 IQKTEFVNVT IQSLMRVTPL RNFFHIPENY QHCKSPLVHC FGELTRKIWH ARNFKGQVSP 180
181 HEFLQAVMKA SKKRFRIGQQ SDPVEFMSWL LNTLHMDLRT SKDASSIIHK CFQGELEVVK 240
241 EFQGNENKEI SRMSFLMLGL DLPPPPLFKD VMEKNIIPQV ALFDLLKKFD GETVTEVVRP 300
301 KLARMRYRVI KSPRYLMFHM VRFKKNNFFK EKNPTLGESV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
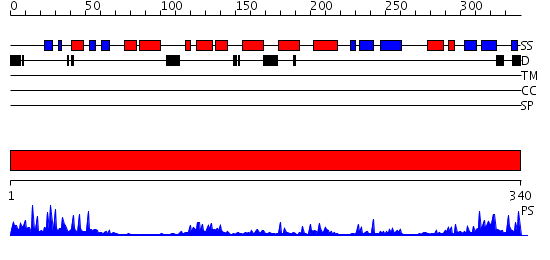
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..340] | 30.69897 | Crystal structure of HAUSP |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)