













| General Information: |
|
| Name(s) found: |
GCSP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1037 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
mitochondrion [IDA] apoplast [IDA] chloroplast envelope [IDA] glycine cleavage complex [ISS] |
| Biological Process: |
glycine decarboxylation via glycine cleavage system
[ISS]
response to cadmium ion [IDA] glycine catabolic process [IMP] |
| Molecular Function: |
protein binding
[IPI]
glycine dehydrogenase (decarboxylating) activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MERARRLAYR GIVKRLVNDT KRHRNAETPH LVPHAPARYV SSLSPFISTP RSVNHTAAFG 60
61 RHQQTRSISV DAVKPSDTFP RRHNSATPDE QTHMAKFCGF DHIDSLIDAT VPKSIRLDSM 120
121 KFSKFDAGLT ESQMIQHMVD LASKNKVFKS FIGMGYYNTH VPTVILRNIM ENPAWYTQYT 180
181 PYQAEISQGR LESLLNFQTV ITDLTGLPMS NASLLDEGTA AAEAMAMCNN ILKGKKKTFV 240
241 IASNCHPQTI DVCKTRADGF DLKVVTSDLK DIDYSSGDVC GVLVQYPGTE GEVLDYAEFV 300
301 KNAHANGVKV VMATDLLALT VLKPPGEFGA DIVVGSAQRF GVPMGYGGPH AAFLATSQEY 360
361 KRMMPGRIIG ISVDSSGKQA LRMAMQTREQ HIRRDKATSN ICTAQALLAN MAAMYAVYHG 420
421 PAGLKSIAQR VHGLAGIFSL GLNKLGVAEV QELPFFDTVK IKCSDAHAIA DAASKSEINL 480
481 RVVDSTTITA SFDETTTLDD VDKLFKVFAS GKPVPFTAES LAPEVQNSIP SSLTRESPYL 540
541 THPIFNMYHT EHELLRYIHK LQSKDLSLCH SMIPLGSCTM KLNATTEMMP VTWPSFTDIH 600
601 PFAPVEQAQG YQEMFENLGD LLCTITGFDS FSLQPNAGAA GEYAGLMVIR AYHMSRGDHH 660
661 RNVCIIPVSA HGTNPASAAM CGMKIITVGT DAKGNINIEE VRKAAEANKD NLAALMVTYP 720
721 STHGVYEEGI DEICNIIHEN GGQVYMDGAN MNAQVGLTSP GFIGADVCHL NLHKTFCIPH 780
781 GGGGPGMGPI GVKNHLAPFL PSHPVIPTGG IPQPEKTAPL GAISAAPWGS ALILPISYTY 840
841 IAMMGSGGLT DASKIAILNA NYMAKRLEKH YPVLFRGVNG TVAHEFIIDL RGFKNTAGIE 900
901 PEDVAKRLMD YGFHGPTMSW PVPGTLMIEP TESESKAELD RFCDALISIR EEIAQIEKGN 960
961 ADVQNNVLKG APHPPSLLMA DTWKKPYSRE YAAFPAPWLR SSKFWPTTGR VDNVYGDRKL1020
1021 VCTLLPEEEQ VAAAVSA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
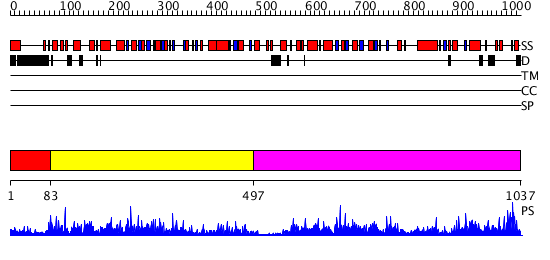
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [83..496] | 73.0 | Crystal structure of glycine decarboxylase (P-protein) of the glycine cleavage system, in apo form | |
| 3 | View Details | [497..1037] | 87.30103 | Crystal structure of glycine decarboxylase (P-protein) of the glycine cleavage system, in apo form |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.81 |
Source: Reynolds et al. (2008)